Abstract
Loss of retinal neurons in adult zebrafish (Danio rerio) induces a robust regenerative response mediated by the reentry of the resident Müller glia into the cell cycle. Upon initiating Müller glia proliferation, their nuclei migrate along the apicobasal axis of the retina in phase with the cell cycle in a process termed interkinetic nuclear migration (INM). We examined the mechanisms governing this cellular process and explored its function in regenerating the adult zebrafish retina. Live-cell imaging revealed that the majority of Müller glia nuclei migrated to the outer nuclear layer (ONL) to divide. These Müller glia formed prominent actin filaments at the rear of nuclei that had migrated to the ONL. Inhibiting actin filament formation or Rho-associated coiled-coil kinase (Rock) activity, which is necessary for phosphorylation of myosin light chain and actin myosin-mediated contraction, disrupted INM with increased numbers of mitotic nuclei remaining in the basal inner nuclear layer, the region where Müller glia typically reside. Double knockdown of Rho-associated coiled-coil kinase 2a (Rock2a) and Rho-associated coiled-coil kinase 2b (Rock2b) similarly disrupted INM and reduced Müller glial cell cycle reentry. In contrast, Rock inhibition immediately before the onset of INM did not affect Müller glia proliferation, but subsequently reduced neuronal progenitor cell proliferation due to early cell cycle exit. Long-term, Rock inhibition increased the generation of mislocalized ganglion/amacrine cells at the expense of rod and cone photoreceptors. In summary, INM is driven by an actin-myosin-mediated process controlled by Rock2a and Rock2b activity, which is required for sufficient proliferation and regeneration of photoreceptors after light damage.
SIGNIFICANCE STATEMENT The human retina does not replace lost or damaged neurons, ultimately causing vision impairment. In contrast, zebrafish are capable of regenerating lost neurons. Understanding the mechanisms that regulate retinal regeneration in these organisms will help to elucidate approaches to stimulate a similar response in humans. In the damaged zebrafish retina, Müller glia dedifferentiate and proliferate to generate neuronal progenitor cells (NPCs) that differentiate into the lost neurons. We show that the nuclei of Müller glia and NPCs migrate apically and basally in phase with the cell cycle. This migration is facilitated by the actin cytoskeleton and Rho-associated coiled-coil kinases (Rocks). We demonstrate that Rock function is required for sufficient proliferation and the regeneration of photoreceptors, likely via regulating nuclear migration.
Keywords: actin cytoskeleton, interkinetic nuclear migration, Müller glia, retinal regeneration, Rho-associated coiled-coil kinases
Introduction
A variety of genetic diseases and environmental insults can lead to damage and loss of retinal neurons in humans, causing permanent vision loss. In contrast to humans, zebrafish can regenerate neurons in various organs including the brain, spinal cord, and eye (Vihtelic and Hyde, 2000; Fausett and Goldman, 2006; Bernardos et al., 2007; Kassen et al., 2007; Kroehne et al., 2011; Dias et al., 2012). In the damaged zebrafish retina, neuronal cell death induces the residing radial glia, the Müller glia, to dedifferentiate and re-enter the cell cycle to produce neuronal progenitor cells (NPCs) that continue to proliferate and subsequently differentiate into and replace the lost neurons (Bernardos et al., 2007; Kassen et al., 2007). Currently, efforts are directed at understanding the molecular mechanisms that govern the regenerative response in the damaged retina (Gorsuch and Hyde, 2014).
During neuroepithelial development, NPC nuclei migrate repeatedly along the apicobasal axis in phase with the cell cycle. As a result of this process, termed interkinetic nuclear migration (INM), NPC nuclei undergo DNA replication (S-phase) in a basal region and perform mitosis at the apical surface of the neuroepithelium (Pearson et al., 2005; Baye and Link, 2007; Xie et al., 2007; Del Bene et al., 2008; Norden et al., 2009; Tsuda et al., 2010; Kosodo et al., 2011; Leung et al., 2011; Lee and Norden, 2013). Disruption of INM is thought to cause premature cell cycle exit of NPCs due to their exposure to different spatial cues along apicobasal Notch-Delta gradients (Murciano et al., 2002; Del Bene et al., 2008). Furthermore, the degree of basal migration during the G1 and S phases was correlated with the decision to either remain in or exit the cell cycle after the next cell division such that more basally located NPC nuclei are more likely to exit the cell cycle than those positioned further medially (Baye and Link, 2007). During INM, nuclei are driven apically in the G2 phase of the cell cycle mainly by actin-myosin-mediated contraction, although microtubules also exert some effect (Del Bene et al., 2008; Norden et al., 2009; Yu et al., 2011). Actin-myosin-mediated contraction requires the phosphorylation of the myosin light chain (MLC), which is mediated by a variety of kinases, including Rho-associated coiled-coil kinases (Rocks) (Amano et al., 1996; Vicente-Manzanares et al., 2009). In the developing retina, Rocks were previously implicated in maintaining apical polarity and facilitating INM (Herder et al., 2013).
Recently, it was suggested that Müller glia also undergo INM during retinal regeneration based on the apical position of phospho-histone 3 (pH3)-positive mitotic nuclei (Nagashima et al., 2013). However, the mechanisms that govern this process in the regenerating retina and the necessity of INM for regeneration are currently unknown. Here, we investigated the mechanisms controlling INM and determined the consequences of its disruption on the regenerative capacity of the light-damaged retina.
Materials and Methods
Fish lines and maintenance.
Adult albino or transgenic Tg[gfap:EGFP]nt11, Tg[atoh7:GFP]rw021 and Tg[rho:Eco.NfsB-EGFP]nt19 zebrafish in an albino background were maintained in the Center for Zebrafish Research at the University of Notre Dame Freiman Life Sciences Center as described previously (Vihtelic and Hyde, 2000). Fish used in these experiments were of either sex at 6–12 months of age and 2–3 cm in length. The protocols used were approved by the University of Notre Dame Animal Care and Use Committee and are in compliance with the statement for the use of animals in vision research by the Association for Research in Vision and Ophthalmology. Adult Tg[gfap:nGFP]mi2004 (13–14 months of age) were kindly provided by the Raymond laboratory.
Light treatment.
The albino and transgenic albino;Tg[gfap:EGFP]nt11, albino;Tg[atoh7:GFP]rw021, albino;Tg[rho:Eco.NfsB-EGFP]nt19, or Tg[gfap:nGFP]mi2004 zebrafish were dark adapted for 14 d and then exposed to constant intense light for 0, 16, 25, 30, 35, 40, 45, 51, 68, or 96 h as described previously (Vihtelic and Hyde, 2000; Kassen et al., 2007). The temperature in the tanks was maintained between 31°C and 33°C. Zebrafish were anesthetized in 1:1000 2-phenoxyethanol and killed in 1:500 2-phenoxyethanol. Eyes were harvested, fixed in either 4% paraformaldehyde (Sigma-Aldrich) in PBS or 9:1 ethanol:formaldehyde (Fisher Scientific) overnight at 4°C and washed three times in PBS or taken through an ethanol gradient and one PBS wash, respectively. Subsequently, eyes were cryoprotected in 30% sucrose in PBS overnight, followed by 1:1 tissue freezing medium to 30% sucrose/PBS overnight and frozen in tissue freezing medium (Triangle Biomedicals) at −80°C.
Drug treatments.
The Rock inhibitor Rockout (25 μm; Calbiochem) or the vehicle control DMSO (1:2000; Life Technologies) were diluted in system water to systemically expose albino;Tg[gfap:EGFP]nt11, albino;Tg[atoh7:GFP]rw021, or albino;Tg[rho:Eco.NfsB-EGFP]nt19 zebrafish starting at 28 h of constant intense light treatment (Yarrow et al., 2005; Harding and Nechiporuk, 2012). To maintain Rockout activity, system water containing the drug was exchanged every 24 h and zebrafish were maintained in a dark incubator set at 30°C until eyes were enucleated at 35, 45, 50, 55, or 72 h after beginning the light treatment. Having previously established that exposure to constant intense light for 24 h was sufficient to induce maximal photoreceptor cell death by 16 h of light damage and a complete regeneration response (Nelson et al., 2013; Conner et al., 2014), we chose to apply Rockout from 28 h of light treatment to prevent any effect it may exert during the period of cell death (Bermel et al., 2009; Koch et al., 2014; Yamamoto et al., 2014). To study the effect of Rockout on retinal regeneration, zebrafish were exposed to constant intense light for 28 h, treated with 25 μm Rockout for 44 h (corresponding to 72 h after beginning the constant light treatment), and subsequently transferred into system water lacking drugs to recover for either 8 or 15 d. In a subset of experiments, the γ-secretase inhibitor R04929097 (750 μm; SelleckBio; Conner et al., 2014) or its vehicle control, DMSO (10%), were intraperitoneally injected into albino;Tg[gfap:EGFP]nt11 zebrafish using a 32 gauge syringe immediately before exposing them to either 25 μm Rockout or DMSO (1:2000) in system water from 28 h of light treatment. Zebrafish were maintained as described above until eyes were harvested at 72 h of light treatment. To maintain R04929097 concentrations at a constant level, intraperitoneal injections of either R04929097 or DMSO were performed every 8 h. The treatment groups were the following: (1) DMSO (1:2000)/DMSO (10%), (2) Rockout/DMSO (10%), (3) DMSO (1:2000)/R04929097, or (4) Rockout/R04929097. In addition, 25 μm cytochalasin D (Tocris Bioscience; Carlier et al., 1986; Becker and Hart, 1999) or its vehicle control, DMSO (1:1000), were injected intraperitoneally into anesthetized zebrafish at 28 h of light treatment using a 32 gauge syringe and maintained in system water in a dark incubator set at 30°C until 35 h of light treatment (Conner et al., 2014). Because the Rho activator CN03 is conjugated to a cell-penetrating moiety, either CN03 (5 μg/ml; Cytoskeleton) or water was intravitreally injected into healthy undamaged albino;Tg[gfap:EGFP]nt11 or albino zebrafish eyes using a Hamilton syringe as described previously (Thummel et al., 2008a). Zebrafish were maintained in a dark incubator at 30°C for 16 h, when albino;Tg[gfap:EGFP]nt11 or albino eyes were collected for immunocytochemical analysis or immunoblotting, respectively.
Morpholino-mediated knockdown in adult zebrafish and embryos.
Lissamine-labeled splice site targeting morpholinos to rock2a (5′-CTCATCTGCACACACAACACAGCAA-3′; GeneTools) and rock2b (5′-GCACACACTCACTCACCAGCTGCAC-3′; GeneTools; Wang et al., 2011) were injected intravitreally together at a final concentration of 0.4 mm each into albino;Tg[gfap:EGFP]nt11 zebrafish and subsequently electroporated as described previously (Thummel et al., 2008a) before initiating constant intense light treatment. The standard control morpholino (0.8 mm) was injected intravitreally and electroporated to monitor potential off-target effects. Eyes were collected at 45 and 72 h of constant light treatment for immunocytochemical analysis and dorsal retinas were isolated at 35 h of light treatment for RNA isolation to confirm successful morpholino-mediated disruption of splicing.
Albino zebrafish embryos were injected at the one- to four-cell stage with lissamine-labeled anti-rock2a, anti-rock2b, or standard control morpholino (Thummel et al., 2008a). Embryos were collected at 24 or 72 h postfertilization for RNA isolation and reverse transcriptase (RT) PCR amplification to test the efficiency of rock2a and rock2b morpholino-mediated disruption of splicing, respectively.
Edu/BrdU pulse chase.
50 μl of 1 mg/ml EdU (Life Technologies) was injected intraperitoneally into anesthetized albino;Tg[gfap:EGFP]nt11 zebrafish using a 32 gauge syringe at 28 h of light treatment and then transferred into system water containing either 1:2000 DMSO or 25 μm Rockout. BrdU (Sigma-Aldrich) was injected intraperitoneally (50 μl) into EdU-treated zebrafish at 44 and 49 h after the start of the light treatment. Eyes were collected for analysis at times corresponding to 45 and 50 h after the start of the light treatment. EdU detection was performed on frozen retinal sections according to manufacturer's guidelines, followed by immunocytochemistry for BrdU (Conner et al., 2014).
To identify newly generated amacrine, ganglion, and bipolar cells at 8 and 15 d of recovery in retinas that were exposed to either DMSO or Rockout from 28–72 h of light treatment, BrdU was intraperitoneally injected into albino zebrafish at 28, 48, 58, 84, 108, and 120 h after the start of the light treatment.
TUNEL assay.
Frozen retinal sections (14 μm) from light-treated albino zebrafish that were exposed to either DMSO (1:2000) or 25 μm Rockout from 28 to either 55 or 72 h after starting the light treatment were analyzed for cell death using the TUNEL assay (Clontech) as described previously (Thummel et al., 2010). Biotin-conjugated dNTPs (Trevigen) and fluorescently labeled streptavidin (1:200; LifeTechnologies) were used to detect TUNEL-positive cells. Retinal sections were subsequently subjected to immunocytochemistry.
Immunocytochemistry, image acquisition, and image analysis.
Immunocytochemistry on frozen sections (14 μm thickness) was performed as described previously (Vihtelic and Hyde, 2000; Kassen et al., 2007; Thummel et al., 2010; Nelson et al., 2012). Primary antibodies used were: rabbit anti-PCNA (1:1000; Abcam), mouse anti-PCNA (1:1000; Sigma-Aldrich), mouse anti- pH3 (1:200; Millipore), rabbit anti-GFP (1:200; Abcam), chicken anti-GFP (1:1000; Abcam), mouse anti-BrdU (1:300; Thermo Scientific), rat anti-BrdU (1:100; Accurate Chemical and Scientific), mouse anti-HuC/D (1:300; Millipore), rabbit anti-PKCα (1:300; Santa Cruz Biotechnology), and rabbit anti-blue (1:500) and rabbit anti-red cone opsins (1:500) (Vihtelic et al., 1999). The pH3 antibody required antigen retrieval before immunocytochemistry as described previously (Raymond et al., 2006; Nelson et al., 2012). Fluorescently tagged secondary antibodies (1:1000; Life Technologies) and the nuclear dye 4′,6-diamidin-2-phenylindol (DAPI, 5 μg/ml; Life Technologies) were applied for 1 h at room temperature, the slides were washed and mounted in Prolong Gold (Life Technologies). Alexa-647 conjugated phalloidin (1:100; Life Technologies) was coapplied with secondary antibodies for 1 h.
Images of the central-dorsal region of the retina were obtained on a Nikon A1 confocal microscope equipped with a 40× plan-fluor oil-immersion objective [numerical aperture (NA), 1.3] and 8–12 μm z-stacks of 1024 × 1024 images were acquired. Cell counts were performed throughout the depth of the z-stack and the length was measured along the horizontal cells using the measurement function in Nikon NIS Elements software to normalize the cell counts to 300 μm length of the imaged retinal section. The position of pH3-positive nuclei was determined in DAPI-labeled sections that distinguished the different retinal nuclear layers. The thickness of the inner nuclear layer (INL) and the distance that pH3-positive cells migrated from the basal limit of the INL were determined using the measurement function in Nikon NIS Elements software (see Fig. 1Y). The pH3-positive Müller glia nuclei were grouped according to their position in: (1) the outer nuclear layer (ONL), (2) the apical part of the INL, or (3) the basal part of the INL, splitting the INL at its half-width. For each retina, the percentage distribution of pH3-positive cells in the different regions was determined and subsequently averaged to give mean distributions for the various experimental groups/conditions.
Figure 1.
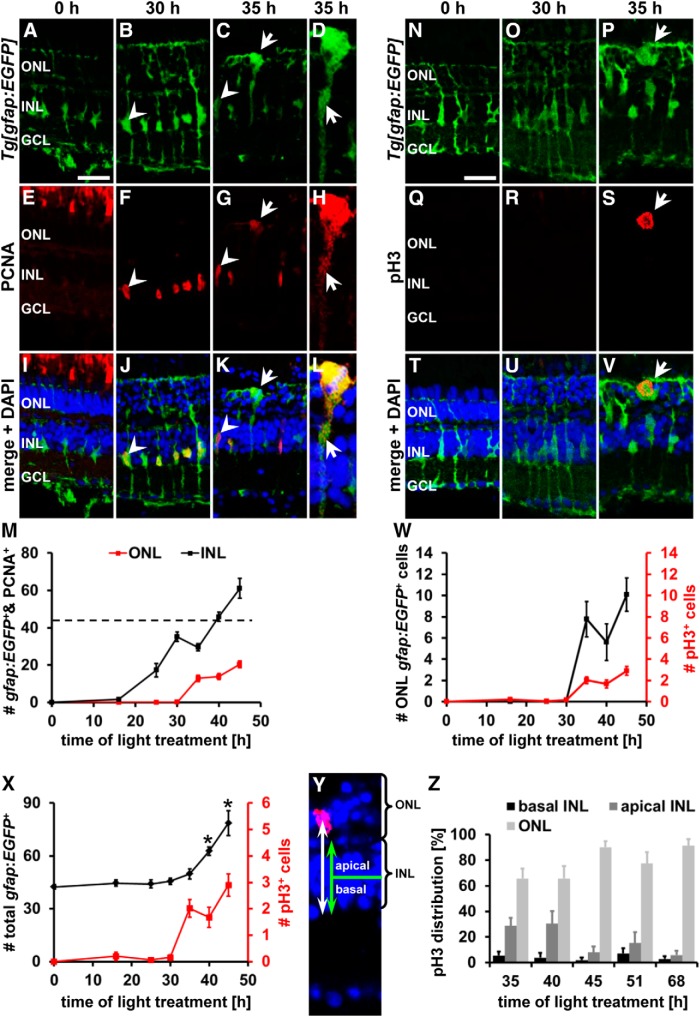
Mitotic Müller glia are present in the ONL beginning at 35 h of light treatment. A–L, N–V, Single z-plane confocal images of albino;Tg[gfap:EGFP]nt11 zebrafish (A–D, I–L, N–P, T–V) colabeled with the proliferation marker PCNA (E–L) or the mitosis marker pH3 (Q–V) and counterstained with the nuclear dye DAPI (I–L, T–V) showing Müller glia in their typical basal position in the INL at 0 and 30 h of light treatment and a subset of Müller glia soma in the ONL at 35 h. Diffuse, cytoplasmic PCNA and the presence of pH3 in ONL Müller glia indicate that these cells were undergoing mitosis (arrow, C, D, G, H, K, L and P, S, V, respectively). Arrowheads indicate EGFP-positive cells containing nuclear PCNA. D, H, L, ONL Müller glia in C, G, and K at higher magnification and adjusted brightness of PCNA to illustrate cytoplasmic PCNA (arrow) in mitotic cells. M, Histogram displaying the number of PCNA and gfap:EGFP double-positive cells in the ONL and INL. Dotted line indicates approximate number of Müller glia in undamaged retinas. Quantification reveals that the onset of Müller glia soma migration to the ONL coincides with the occurrence of pH3 at 35 h (W) and that the total number of gfap:EGFP-positive cells significantly increased at 40 and 45 h, immediately after the onset of Müller glia nuclear migration (X). Y, Distance that pH3-positive Müller glia nuclei migrated from the basal limit of the INL (white arrow) and the INL width (green arrow) were measured on confocal sections labeled with DAPI (blue) and pH3 (red). pH3-positive nuclei in the basal retina represent Müller glia in their typical position, whereas those in the apical portion had migrated. Z, Histogram of the distribution of pH3-positive nuclei in the retina during the light treatment time course. GCL, Ganglion cell layer. Data are shown as mean ± SE, n ≥ 10, *p < 0.05, ANOVA, Bonferroni post hoc test compared with 30 h of light treatment. Scale bars, 20 μm in A–C, E–G, I–K, and N–V.
Statistical analysis.
Data were subjected to Student's t test for statistical analysis of single comparisons between the control and the treatment group. Multiple data groups were tested for statistical significance using one-way ANOVA followed by a Bonferroni's or Tukey's post hoc test. The test used in each experiment, the n value, and the level of statistical significance (p value) are indicated in the corresponding figure legends. In the corresponding Results section, the statistical values for each experiment were also included; however, the statistical test was only mentioned if a test other than Student's t test was applied.
Immunoblot.
Immunoblotting was performed as described previously (Kassen et al., 2007). Briefly, dorsal or whole retinas were collected from (1) light-damaged and Rockout-treated, (2) light-damaged and DMSO-treated, (3) undamaged and CN03-injected, or (4) undamaged H2O-injected zebrafish. The retinas were homogenized in lysis buffer containing PBS supplemented with Triton X-100, proteinase inhibitors (Roche), and phosphatase inhibitor II and III (Sigma-Aldrich). Then, 20 μg of protein was electrophoresed through 4–20% Tris-glycine gels (Life Technologies) and transferred onto PVDF membranes (GE Healthcare). Membranes were blocked in 5% milk in Tris-buffered saline overnight and probed with antibodies to phospho (Thr 18/Ser 19)-MLC (1:250; Cell Signaling Technology) or actin (loading control, 1:1000; Sigma-Aldrich) for 4 h at room temperature. HRP-conjugated secondary antibodies were applied for 1 h at room temperature and membranes were developed using Pierce ECL Western Blotting substrate (Fisher Scientific). Scanning laser densitometry was performed on Western blots that compared phospho (Thr 18/Ser 19)-MLC expression levels between Rockout- and DMSO-treated retinas using ImageJ and were normalized to the corresponding actin expression levels.
Retinal culture/live-cell imaging.
Adult (6–12 months of age) albino;Tg[gfap:EGFP]nt11 zebrafish were exposed to constant intense light for 35 h, the eyes were enucleated, and the sclera and lens removed to obtain the retina. The retina was cut from the optic stalk toward the anterior and posterior sites and the dorsal portion was mounted on Cell-Tak-coated (70 μg/ml diluted in 0.1 mm NaHCO3; BD Biosciences) Fluoro-dishes (World Precision Instruments) with the ganglion cells facing the glass coverslip. Retinas were overlaid with 1% agarose dissolved in MEM and cultured in MEM without phenol red containing penicillin/streptomycin (Thermo Scientific), 25% horse serum, 25% Hank's balanced salt solution supplemented with 1 mm CaCl2 and 1 mm MgCl2 (adjusted from Surzenko et al., 2013), and incubated in a 5% CO2/air environment at 32°C. The materials required for culturing retinal explants were obtained from LifeTechnologies unless otherwise stated. Because Tg[gfap:nGFP]mi2004 zebrafish (13–14 months of age) were pigmented, they were exposed to constant intense light for 40 h to ensure that their retinas had been readily damaged. Cultures from Tg[gfap:nGFP]mi2004 zebrafish were prepared as described above.
Live-cell imaging was performed using an inverted Nikon A1 Multiphoton microscope equipped with a 40× Apo long distance water-immersion objective (NA 1.15). Retinal cultures were maintained at a temperature of 27–29°C in a gas chamber supplied with 5% CO2/air for the duration of the experiment starting 48 h after the onset of the light treatment. The EGFP was excited with a Mai Tai IR laser (Spectraphysics) set at 910 nm and 512 × 512 images were acquired at a zoom of two. For cultures from Tg[gfap:nGFP]mi2004 zebrafish, images were also acquired at a zoom of two, but were cropped to a field of view of 340 × 340 pixels to increase the z-stack acquisition speed. Time-lapse imaging was performed continuously for 5–6 h, recording z-stacks at 0.7 μm z-step size that extend from the ganglion to the photoreceptor cell layer. Relative z-intensity correction was applied to compensate for loss of z-depth intensity. 3D reconstructions were prepared for a subregion using Nikon elements software. Data analysis was performed on three independent retinas for both albino;Tg[gfap:EGFP]nt11 and Tg[gfap:nGFP]mi2004 cultures. To determine the velocities of apical and basal nuclear migration, maximum x–z or y–z projections were prepared from cropped subregions that contained a dividing Tg[gfap:nGFP]mi2004-positive nucleus using Nikon elements software. Because only nuclei are labeled in Tg[gfap:nGFP]mi2004 zebrafish, the basal limit of the retina could not be visualized and could therefore not serve as reference to measure distances that nuclei had migrated. Therefore, to obtain a reference point, we identified a Tg[gfap:nGFP]mi2004-positive nucleus positioned in the basal INL in the maximum projection that did not migrate throughout the imaging period. The distance that a nucleus migrated was measured relative to this reference nucleus using Nikon elements software and plotted against the time using Microsoft Excel. To determine the velocities of apical (va) and basal (vb) nuclear migration, linear regression curves (y = mx + c) were fitted in which the slope “m” represents the velocity (dx/dt). To determine va, a linear regression curve was fitted to the period of nuclear movement before the nuclear envelope broke down; for vb, linear regression analysis was performed on the initial period of rapid basal movement (see Fig. 2I).
Figure 2.
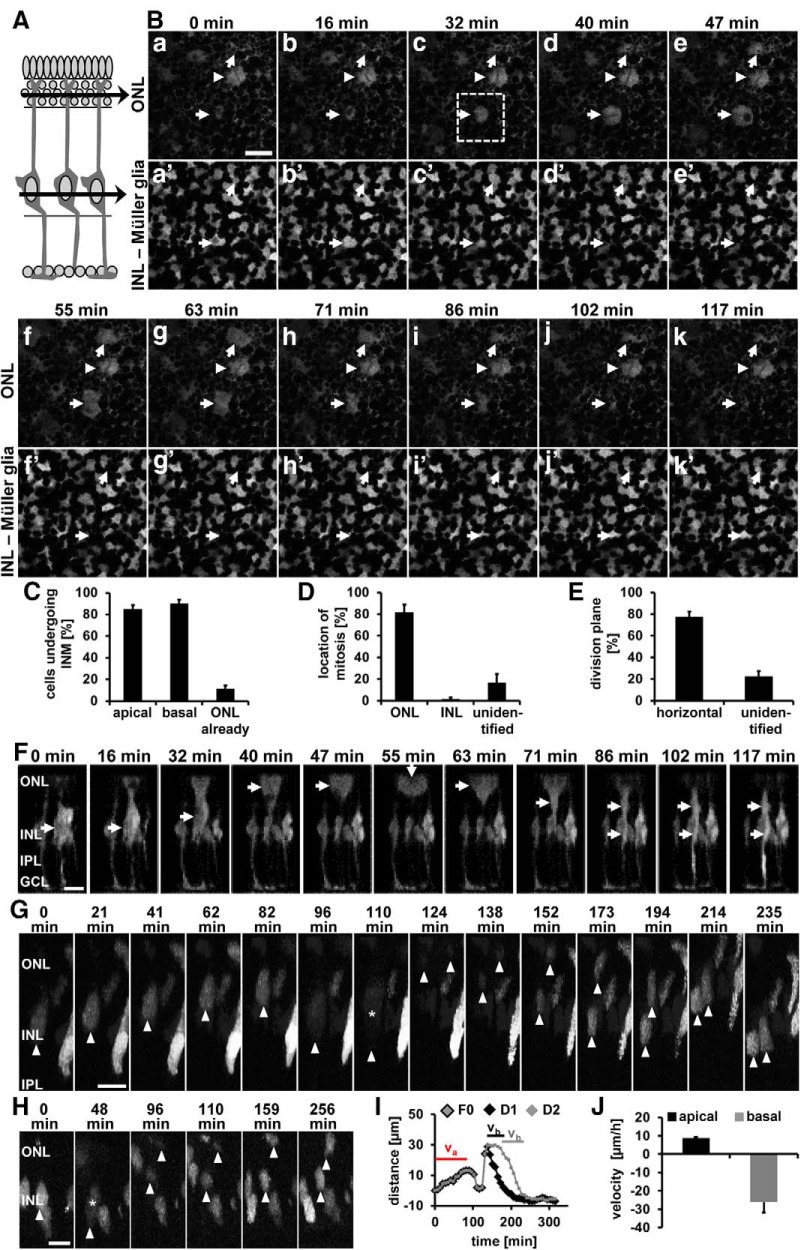
Live-cell imaging of INM in adult zebrafish retinal explants. A, Schematic of a retinal cross-section. The black arrows indicate the z-planes at the level of the ONL (Ba–Bk) and Müller glia soma (Ba′–Bk′). B, Time-lapse image series of retinal explant cultures from adult albino Tg[gfap:EGFP]nt11 zebrafish at 48 h of light treatment acquired by multiphoton microscopy showing that Müller glia divide in the ONL (arrows) and that their division plane is horizontal to the surface of the epithelium. Arrowheads indicate a Müller glia present in the ONL at the onset of image acquisition that displayed hallmarks of mitosis but did not divide during the 2 h recording. Scale bar, 20 μm. C–E, Histograms depicting the percentage of cells undergoing INM (C), the location of mitosis (D), and the division plane (E). F, Time-lapse image series of a 3D reconstruction of the z-series for the cell surrounded by a square in Bc. Arrows indicate the position of the cell soma as it migrates apically to the ONL and returns basally. Scale bar, 20 μm. G, H, Time-lapse image series of a 3D reconstruction of retinal cultures from adult Tg[gfap:nEGFP]mi2004 zebrafish giving an example of cells dividing horizontally (G) or vertically (H). Arrowheads point to the basal region of the nucleus or the soma after nuclear membrane breakdown. Stars indicate the time point of nuclear membrane breakdown. Scale bars in G, H, 10 μm. I, Graph depicting the distance the nucleus (F0) displayed in G and its daughter nuclei (D1, D2) traveled in relation to the basal INL position throughout the time lapse. Bars above the trace indicate the time period used to calculate the velocities of apical (va) and basal migration (vb). J, Histogram displaying the velocities of apical and basal nuclear migration. Images are representative from three independent retinal cultures. Data are shown as mean ± SE, nTg[gfap:EGFP] = 51 cells, nTg[gfap:nGFP] = 80 cells from three different retinal explants each. GCL, Ganglion cell layer.
Quantitative real-time PCR and RT-PCR.
RNA was isolated from dorsal retinas obtained from light-damaged albino zebrafish (0, 16, 35, 51, 68, and 96 h) using TRIzol (Life Technologies) according to the manufacturer's instructions and stored at −80°C. cDNA was synthesized using Superscript III First Strand synthesis kit (Life Technologies) and subjected to quantitative real-time PCR as described previously (Nelson et al., 2012). Primers used in this study were as follows: rock1 (F: 5′ GAGGTTTTTGCAGAGGACAG, R: 5′ CAGTCTCCTCGATCAATGTC), rock2a (F: 5′ GAAAACCGGCTGAGACAACT, R: 5′ TAAGGCCGGAAAGTCCAGAT), rock2b (F: 5′ GGGTGAGAAAACTAGAGACG, R: 5′ CTGGATGGTTCAGATCAAGG), 18 S (F: 5′ AATTGACGGAAGGGCACCAC, R: 5′ CTAAGAACGGCCATGCACCA). Quantitative real-time PCR was performed on a minimum of three biological replicates, as well as three technical replicates for rock1, rock2a, and rock2b transcripts.
To confirm that rock2a and rock2b morpholinos successfully prevented splicing of the corresponding mRNA, cDNA was synthesized from isolated RNA as described above. The following primers were used for reverse transcriptase PCR: rock2a: (F: 5′ TCTGGACAAGTCCGGGGTA, R: 5′ AGATCTCCACCTGGCATGTA), rock2b: (F: 5′ GGATTCAGTAACTGCCCTTG, R: 5′ GAGTTTGAGGTGTCCATTGC). The PCR products were gel extracted using QIAquick gel extraction kit (Qiagen), subcloned into pCR4-TOPO vectors (Life Technologies) according to the manufacturer's guidelines, and sequenced at the Notre Dame Genomics facility to confirm their identity.
Results
Mitotic Müller glia and NPCs are localized in the ONL
Recently, mitotic Müller glia nuclei were observed in the ONL of light-damaged zebrafish retinas after brief exposure to high-intensity light, which suggests that proliferating Müller glia undergo INM similar to NPCs during retinal development (Pearson et al., 2005; Baye and Link, 2007; Del Bene et al., 2008; Norden et al., 2009; Nagashima et al., 2013). To investigate the mechanisms that regulate INM during retinal regeneration, we initially confirmed that Müller glia undergo INM in dark-adapted zebrafish that were exposed to constant intense light that causes photoreceptor cell death (Vihtelic and Hyde, 2000; Vihtelic et al., 2006). Retinal sections from light-damaged albino;Tg[gfap:EGFP]nt11 zebrafish, which express EGFP specifically in all Müller glia under the control of the glial fibrillary acidic protein (gfap) promoter (Kassen et al., 2007), were colabeled with the proliferation marker proliferating cell nuclear antigen (PCNA). PCNA-positive Müller glia were first observed at 25 h of light treatment (Fig. 1M). At this time point and at 30 h of light treatment (Fig. 1F,J,M), PCNA-positive Müller glia were located in the basal region of the INL, where quiescent Müller glia typically reside in the undamaged retina (Fig. 1A,B,E,F,I,J,M). In contrast, at 35 h of light treatment, a subset of Müller glia had migrated apically into the ONL (Fig. 1C,G,K,M), the equivalent region where NPCs undergo mitosis during retinal development. Compared with their typical stellate appearance in the INL (Fig. 1A,B), these ONL Müller glia were often increased in size and rounded (Fig. 1C), which are both hallmarks of dividing cells (Meyer et al., 2011; Spear and Erickson, 2012). Moreover, Müller glia positioned in the ONL were characterized by diffuse labeling of PCNA in the cytoplasm that was excluded from DAPI-labeled mitotic chromatin structures (Fig. 1C,D,G,H,K,L, arrows) in contrast to their INL counterparts that displayed nuclear PCNA (Fig. 1G,K, arrowheads). Exclusion of PCNA from the nucleus occurs during mitosis after nuclear membrane breakdown (Hutchison and Kill, 1989; Leung et al., 2011). At later stages during the light damage time course (40 and 45 h), gfap:EGFP-positive cells, representing either Müller glia or their derived NPCs, were also observed in the ONL (Fig. 1M). The timing between the first upregulation of PCNA in the Müller glia at 25 h of light treatment and the onset of INM at 35 h suggests that the combined length of the G1, S, and G2 phases of proliferating Müller glia is ∼10 h. This is in good agreement with the approximate time that is required to complete the G1, S, and G2 phases during retinal development (Leung et al., 2011). Subsequent to Müller glia migration to the ONL at 35 h of light treatment, the number of PCNA-positive cells in the INL steadily increased above the number of gfap:EGFP cells that are observed in the undamaged retina (Fig. 1M, dotted horizontal line), indicating that the onset of INM correlates with mitosis and the generation of NPCs. In support, the time when Müller glia migration to the ONL was first observed coincided with the occurrence of a few PCNA doublets in the INL indicating that cell division had taken place in some Müller glia (data not shown and Nelson et al., 2012). Together, the morphological changes of ONL Müller glia, their cytoplasmic localization of PCNA, the mitotic chromatin structures, and the increase in proliferating INL cells above the number of Müller glia present in the undamaged retina strongly suggest that Müller glia migrated to the apical retina to undergo mitosis starting at 35 h of light treatment.
To confirm that Müller glia migrated to the ONL to undergo mitosis, we immunocytochemically stained light-damaged retinal sections from albino;Tg[gfap:EGFP]nt11 zebrafish with an antibody to pH3 that labels cells in the late stage of G2 and during early phases of mitosis (Hendzel et al., 1997). In both undamaged and 30 h light-damaged retinas, gfap:EGFP-positive Müller glia in the INL did not colabel with pH3 (Fig. 1N,O,Q,R,T,U,W). At 35 h, pH3 colocalized with DAPI-positive mitotic chromatin structures in EGFP-positive cells in the ONL (Fig. 1P,S,V) and persisted at later time points (40 and 45 h of light treatment; Fig. 1W), confirming that cells divided in the ONL in our light damage model. In support of cell division occurring, the total number of gfap:EGFP-positive cells (sum of those in the INL and ONL) significantly increased from 45.6 ± 1.8 cells/300 μm at 30 h (time point before mitosis, n = 14) to 62.8 ± 2.3 cells/300 μm (Fig. 1X; n = 11, ANOVA, p = 0.0003, Fcrit = 3.33; Bonferroni test between 30 and 40 h, p < 0.05) and 78.7 ± 7.1 cells/300 μm (n = 12, Bonferroni test between 30 and 45 h, p < 0.05) at 40 and 45 h, respectively. This indicates that Müller glia exhibited INM to the ONL to undergo cell division that generated NPCs.
In addition to pH3-positive ONL Müller glia, we also observed a subset of pH3-positive cells in the apical and basal INL at 35 h. To determine the frequency of pH3-positive Müller glia localizing to the ONL versus the apical or basal INL during retinal regeneration, we labeled retinal sections with the nuclear dye DAPI to identify the boundaries of the INL and ONL. The percentage of pH3-positive Müller glia present in the basal and apical halves of the INL and the ONL were determined (Fig. 1Y). At 35 h of light treatment 65.8 ± 7.8% of the pH3-positive Müller glia were localized in the ONL (n = 13), whereas 5.4 ± 3.1% and 28.8 ± 6.3% were observed in the basal and apical INL, respectively (Fig. 1Z). A similar distribution was observed at 40 h of light treatment (Fig. 1Z; ONL: 65.8 ± 9.5%, apical INL 30.4 ± 9.8% and basal INL 3.8 ± 3.8%, n = 13). However, 90.1 ± 4.6% of the pH3-positive, gfap:EGFP-positive cells (Fig. 1Z; n = 13) were present in the ONL at 45 h of constant exposure to intense light, a time point when the first NPCs undergo mitosis, as evidenced by the occurrence of four PCNA-positive cells in some clusters (Fig. 1Z and data not shown). This predominant ONL distribution of pH3-positive cells is maintained at subsequent time points when NPCs divide (Fig. 1Z; 51 h: 77.5 ± 8.7%, n = 12; 68 h: 91.3 ± 5.3%, n = 10). These data suggest that both Müller glia and NPCs underwent INM to the ONL.
Live-cell imaging confirms that Müller glia/NPCs undergo INM
The position of pH3-positive nuclei along the apicobasal axis of the retina suggests that Müller glia undergo INM in the adult regenerating retina. To confirm this hypothesis, we performed live-cell imaging of adult retinal cultures from albino;Tg[gfap:EGFP]nt11 zebrafish. Retinas isolated at 35 h of light treatment were cultured as whole mounts for 10–12 h to recover from the acute damage suffered before z-stack time-lapse imaging was performed through the retinal thickness by multiphoton microscopy. Single images from two focal planes, the basal INL, where Müller glia reside, and the ONL, were chosen as indicated (Fig. 2A). Cell bodies of a subset of gfap:EGFP-positive cells that were located in the INL at the onset of imaging (arrows, Fig. 2Ba′, Bb′) decreased in size over time (arrows, Fig. 2Bc′–Be′), whereas gfap:EGFP-positive soma became present in the ONL (arrows, Fig. 2Bc–Be). These ONL gfap:EGFP-positive soma had a round morphology, characteristic for cells undergoing mitosis (Meyer et al., 2011; Spear and Erickson, 2012). After 55 and 63 min of imaging, two gfap:EGFP-positive soma in the ONL showed clear signs of cell division (arrows, Fig. 2Bf, Bg). Typically, those dividing cells pulled apart and the cleavage plane became visible (arrow pointing to cleavage plane, Fig. 2Bf, Bg) before they collapsed and decreased their size in the ONL (Fig. 2Bh–Bk) while returning to the INL (Fig. 2Bh′–Bk′). Of the cells observed, 85.3 ± 3.6% and 90.3 ± 3.6% of the gfap:EGFP-positive soma migrated apically and basally, respectively, during the time of recording (Fig. 2C; 51 dividing cells, 3 independent retinal cultures). A subset of cells were already positioned in the ONL (Fig. 2C; 11.5 ± 2.9%) at the onset of imaging. Occasionally, we observed a gfap:EGFP-positive cell in the ONL that exhibited a round morphology typical for being in a mitotic state, but remained there for several hours before cell division took place. For example, one ONL cell (arrowhead, Fig 2Ba–Bk) took >6 h to divide (data not shown). The majority of cells divided in the ONL (Fig. 2D, 81.8 ± 6.9%), whereas only 1.4 ± 1.4% underwent cell division in the INL where Müller glia typically reside. The remaining gfap:EGFP-positive cells (Fig. 2D; unidentified: 16.8 ± 8.0%) migrated first apically and then basally, but the location of the cell division was not identifiable. The cleavage plane of 77.4 ± 4.8% of the ONL gfap:EGFP-positive cells (41 of 51 observed) was vertical to the apical surface of the retina (Fig. 2Bf, E, arrows are aligned with cleavage plane), which is consistent with the horizontal alignment of the spindle apparatus and anaphase chromatin along the apical surface during retinal development (Horne-Badovinac et al., 2001; Cayouette and Raff, 2003). Therefore, this type of cell division will be referred to as “horizontal” to conform to nomenclature used during retinal development. The division plane of the residual 22.6 ± 4.8% of the ONL gfap:EGFP-positive cells could not be convincingly identified (Fig. 2E). 3D reconstructions of z-stack images through the depth of the retina further confirmed that gfap:EGFP-positive soma migrated to the ONL to divide horizontally before migrating basally into the INL (Fig. 2F).
To further characterize the migratory and division pattern of Müller glia/NPC nuclei, we prepared retinal cultures from Tg[gfap:nGFP]mi2004 zebrafish, which localize GFP in the nucleus of activated Müller glia/NPCs (Bernardos et al., 2007; Nagashima et al., 2013). Similar to albino;Tg[gfap:EGFP]nt11 retinal cultures, 82.46 ± 3.39% of dividing Tg[gfap:nGFP]mi2004 nuclei (80 dividing cells, 3 independent retinal explants) migrated apically into either the upper INL or ONL to undergo mitosis (Fig. 2G), whereas 16.5 ± 3.8% already resided in the ONL at the onset of the time lapse. Although the majority of nuclei divided in the ONL (85.6 ± 4.3%), 1.1 ± 1.1% and 3.8 ± 0.3% of nuclei divided in the basal and apical INL, respectively. The position of the remaining nuclei that divided vertically (9.5 ± 3.6%) could not be classified as either ONL or INL because one nucleus was typically observed in the ONL and the other in the INL during mitosis (Fig. 2H). Regardless of the location of the nuclear division, the predominant orientation of the division plane was horizontal (Fig. 2G; 90.5 ± 3.6%).
Nuclei migrated, on average, 11.8 ± 1.2 μm apically (Fig. 2I) with a velocity of 8.8 ± 0.5 μm/h (Fig. 2J; R2 = 0.89 ± 0.020) before the nuclear envelope broke down as evidenced by the distribution of GFP throughout the whole cell (Fig. 2G, 110 min; H, 48 min). Upon nuclear envelope breakdown, the position of chromatin could not be measured and thus it was not possible to determine the velocity of the second phase of apical INM. During this second phase of apical INM, the chromatin migrated an additional 11.4 ± 1.1 μm based on the difference between the last position measured before nuclear envelope breakdown and the first position after nuclei became visible after mitosis. We attempted to label retinal cultures with Hoechst to monitor the migration of nuclei after nuclear membrane breakdown and to determine the velocity of the second phase of apical INM. However, the long-term presence of Hoechst in retinal cells, in conjunction with multiphoton imaging, resulted in toxicity and stalling of the cell cycle (data not shown). Mitosis lasted on average 38.1 ± 5.2 min before 61.4 ± 15.9% of cells migrated 26.6 ± 2.7 μm basally into the INL (Fig. 2I) with an average velocity of −26.2 ± 5.7 μm/h (Fig. 2J; R2 = 0.94 ± 0.0087), suggesting that this movement might be driven by an active mechanism. Live-cell imaging of retinal whole-mount cultures convincingly demonstrated that Müller glia/NPCs underwent INM to primarily divide horizontally in the ONL.
Actin cytoskeleton mediates INM during retinal regeneration
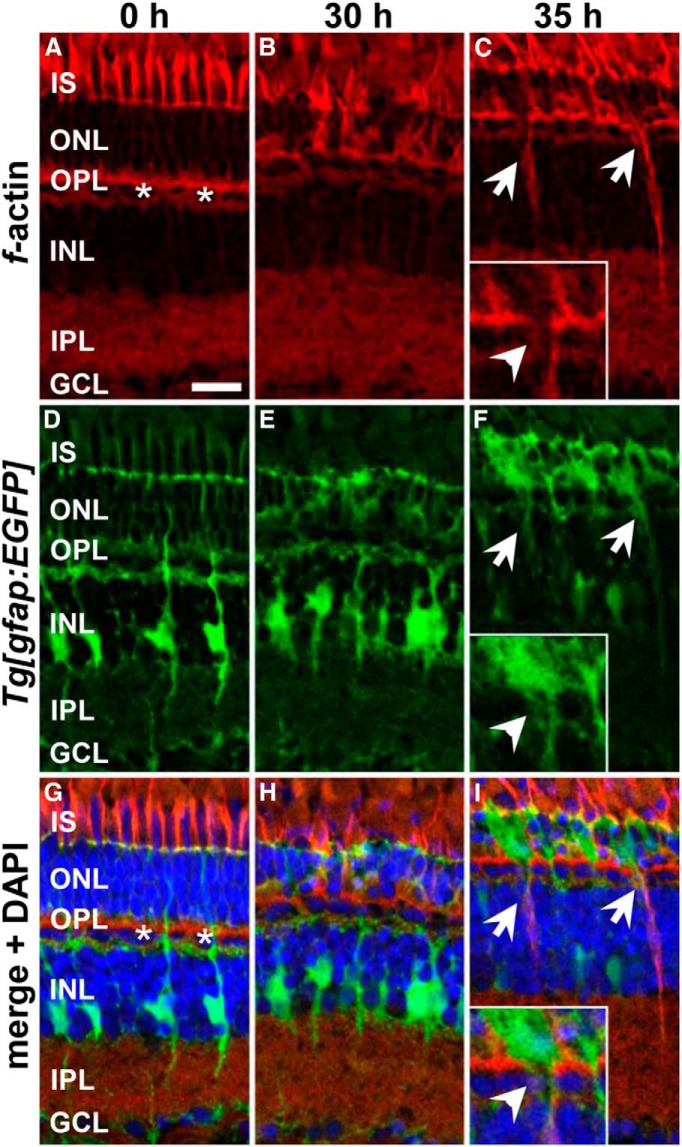
During retinal development, it was shown that filamentous actin (f-actin) accumulates at the rear of migrating NPC nuclei and that actin-myosin-mediated contraction facilitates INM (Norden et al., 2009). If the actin cytoskeleton were critical in mediating INM, then we might expect to observe a similar accumulation of f-actin in Müller glia at the onset of INM. Therefore, we labeled retinal sections with fluorescently tagged phalloidin, which specifically binds f-actin. In undamaged retinas, phalloidin labeled the photoreceptor inner segments, the outer plexiform layer and the inner plexiform layer (IPL) (Fig. 3A,D,G) as described previously (Lewis et al., 1995; Hale et al., 1996). Moreover, the nuclei of the horizontal cell layer were strongly surrounded by a network of f-actin (Fig. 3A,G, asterisks) and cells/nuclei in the ONL and INL were weakly outlined (Fig. 3A,G). At 30 h of light treatment, when Müller glia had re-entered the cell cycle, a similar pattern was observed except that the ONL and the photoreceptor inner segments displayed a high degree of disorganization resulting from the loss of photoreceptors (Fig. 3B,E,H). At 35 h of light treatment, when Müller glia INM had initiated, f-actin strongly localized to the processes of gfap:EGFP-positive Müller glia that were positioned in the ONL (Fig. 3C,F,I). Moreover, the actin filaments surrounding the horizontal cell layer were discontinuous at sites where Müller glia were migrating into the ONL (inset, Fig. 3C,F,I). These data showed a clear correlation among the onset of actin filament formation in Müller glia, the structural changes of f-actin lining the horizontal cell layer, and the onset of INM, suggesting that this cytoskeletal component facilitated nuclear migration.
Figure 3.
Formation of f-actin at the rear of migrating Müller glia at 35 h of light treatment. Single z-plane confocal images of phalloidin-labeled f-actin (A–C) in retinal sections from albino;Tg[gfap:EGFP]nt11 zebrafish (D–F) that were either nondamaged (0 h; A, D, G) or light treated for 30 (B, E, H) or 35 h (C, F, I). Merged images that were counterstained with the nuclear dye DAPI are displayed in G–I. Asterisks indicate horizontal cells that are surrounded by f-actin. Arrows indicate f-actin in the rear of ONL Müller glia. Inset (C, F, I) marked discontinuous f-actin labeling lining the horizontal cell layer at sites where Müller glia migrate into the ONL (arrowhead). GCL, Ganglion cell layer; IS, inner segments. Scale bar, 15 μm in A–I.
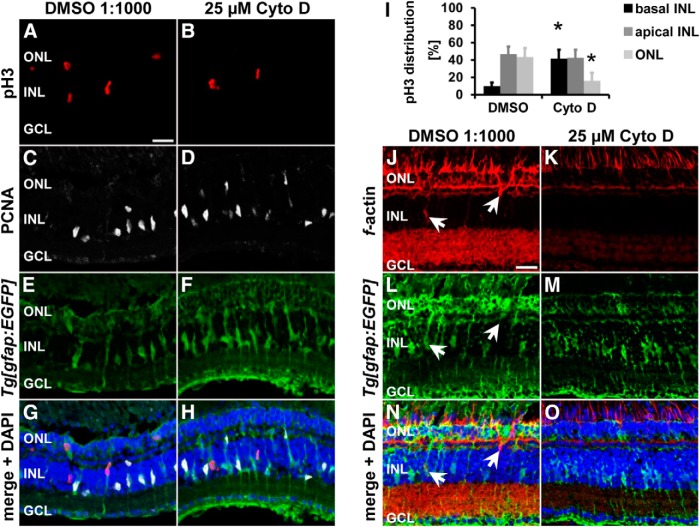
To test functionally whether the actin cytoskeleton mediates INM, we intraperitoneally injected either the actin polymerization inhibitor cytochalasin D (25 μm; Carlier et al., 1986; Becker and Hart, 1999) or the vehicle control DMSO (1:1000) at 28 h of light treatment, before the onset of INM. The positions of pH3-positive nuclei were analyzed to determine whether inhibiting f-actin formation disrupts nuclear migration. Cytochalasin D significantly increased the percentage of pH3-positive nuclei in the basal INL region, where quiescent Müller glia typically reside (Fig. 4A–I; cytochalasin D: 41.7 ± 10.3%, n = 11; DMSO: 10.0 ± 4.3%, n = 13, p = 0.003) at the expense of those in the ONL relative to DMSO vehicle controls at 35 h of light treatment (Fig. 4A–I; cytochalasin D: 15.9 ± 9.5%, n = 11; DMSO: 43.4 ± 10.3%, n = 13, p = 0.033). Phalloidin staining showed reduced levels of labeled actin filaments in retinas exposed to cytochalasin D compared with those treated with DMSO (Fig. 4J–O). In addition, only 4 of 11 cytochalasin D-treated retinas contained actin filaments at the rear of Müller glia, whereas 7 out 8 DMSO-exposed retinas exhibited phalloidin-labeled actin filaments in Müller glia, which confirmed that cytochalasin D reduced actin filament formation. Together, both the presence of actin filaments at the rear of migrating Müller glia nuclei at the onset of INM and the increase in basally located mitotic nuclei after cytochalasin D exposure indicated that the actin cytoskeleton drove Müller glia INM during retinal regeneration.
Figure 4.
Cytochalasin D alters the localization of pH3-positive nuclei at 35 h of light treatment. A–H, J–O, Single z-plane confocal images of albino;Tg[gfap:EGFP]nt11 zebrafish that were intraperitoneally injected with either DMSO (A, C, E, G, J, L, N) or 25 μm cytochalasin D (CytoD; B, D, F, H, K, M, O) and immunocytochemically labeled with pH3 (A, B, G, H) and PCNA (C, D, G, H) or phalloidin (J, K, N, O) and counterstained with the nuclear dye DAPI (G, H, N, O). Exposure to cytochalasin D altered the position of pH3-positive Müller glia (A, B, G, H), but did not affect the number of PCNA-positive cells (C, D, G, H) at 35 h of light treatment. I, Histogram depicting the distribution of pH3-positive nuclei in the ONL and apical and basal INL in DMSO- and cytochalasin D-treated zebrafish at 35 h of light treatment. J–O) Reduced intensities of phalloidin-labeling were observed in cytochalasin D-treated retinas compared with those that received DMSO. Arrows indicate f-actin at the rear of migrating Müller glia nuclei. Data are shown as mean ± SE, n ≥ 11, Student's t test, *p < 0.05 between corresponding regions in DMSO and cytochalasin D-treated samples. GCL, Ganglion cell layer. Scale bar, 20 μm in A–H and J–O.
Rock2a and rock2b mRNA levels are not significantly upregulated in the light-damaged retina
Actin-myosin-mediated contraction is regulated through phosphorylation of MLC, which can be mediated by various kinases that include Rocks, MLC kinases, and citron kinase (Vicente-Manzanares et al., 2009). Recently, Rock2a was implicated in controlling the position of mitotic nuclei during retinal development (Herder et al., 2013). To determine whether Rocks might play a role in mediating INM during retinal regeneration, we determined the temporal expression pattern of the different members of the rock gene family (rock1, rock2a, and rock2b) in the light-damaged zebrafish retina using qRT-PCR. Baseline levels of rock1 mRNA were observed throughout the light damage time course (Fig. 5; ANOVA, p = 0.94). In contrast, rock2a and rock2b mRNA levels were upregulated relative to undamaged control retinas (0 h) at 16 and 35 h of light treatment, although this increase was not statistically significant (Fig. 5; ANOVA, rock2a: p = 0.26; rock2b: p = 0.56). Although the mRNA levels were not significantly increased, small changes in mRNA levels could be amplified at the translational level. Therefore, the small changes in rock2a and rock2b mRNA levels at 35 h of light treatment, when Müller glia nuclear migration was first observed, might result in significant increases in Rock2a and Rock2b protein that might play a role in INM.
Figure 5.
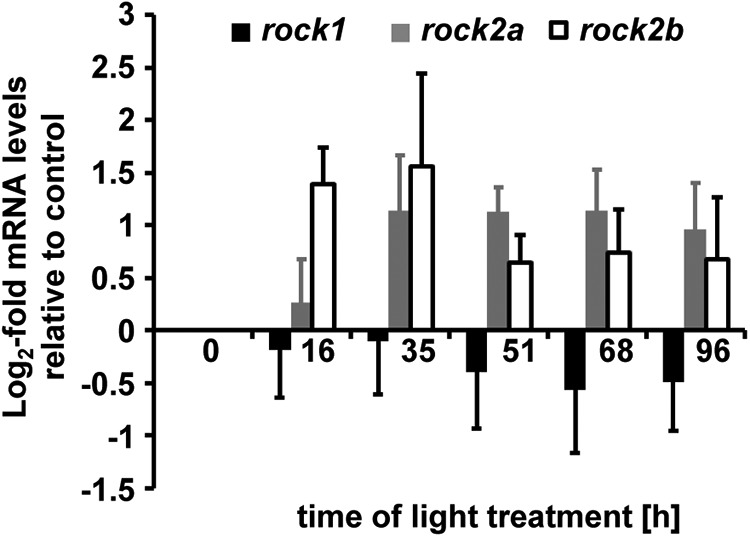
Temporal mRNA expression patterns of rocks. mRNA was isolated from dark-adapted undamaged (0 h) and light-damaged retinas (16–96 h) and expression levels were analyzed by qRT-PCR. Both rock2a and rock2b expression increased in light-damaged retinas relative to undamaged controls (0 h), although not significantly. In contrast, rock1 mRNA remained relatively constant. Data are shown as mean ± SE, n ≥ 3, ANOVA, p > 0.05.
Rock2a and Rock2b are required for apical migration in light-damaged retinas
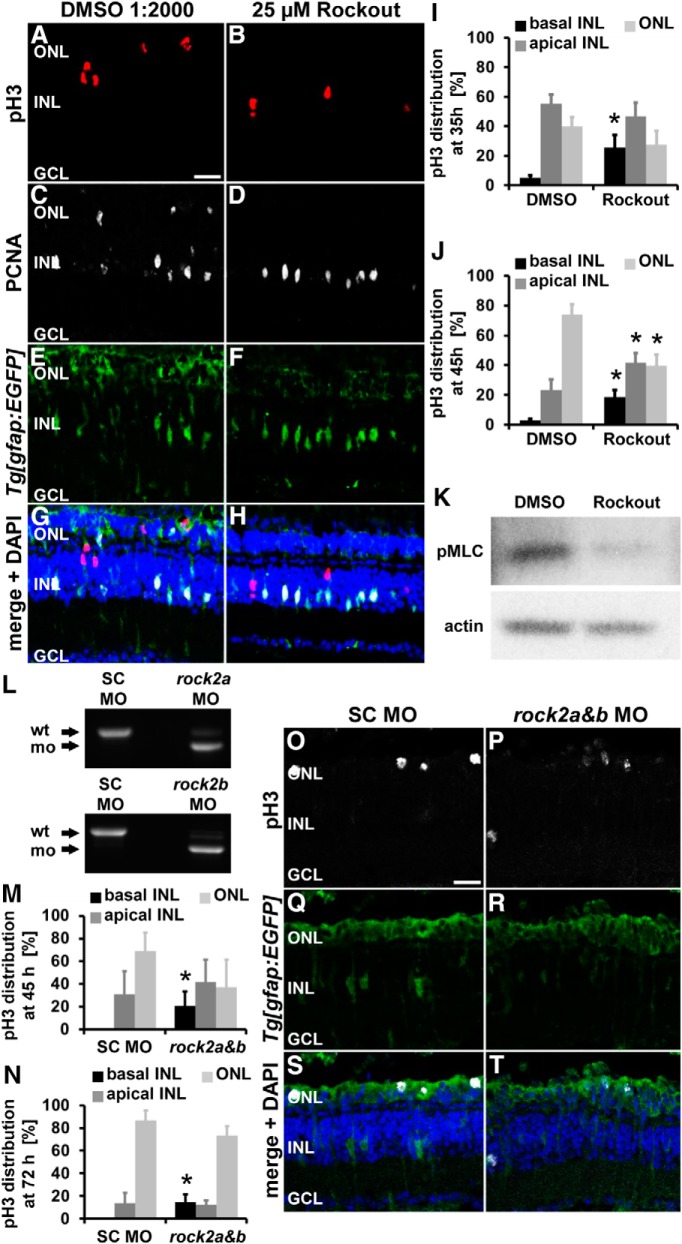
Because Rock2 was implicated previously in mediating INM in the developing retina, we used the Rock inhibitor Rockout to determine whether Rocks are required for actin-myosin-mediated contraction during INM in the regenerating retina. albino;Tg[gfap:EGFP]nt11 zebrafish were systemically exposed to 25 μm Rockout or the vehicle control DMSO (1:2000) starting at 28 h of light treatment and the position of mitotic Müller glia (pH3-positive) were assessed after 35 h of light treatment. Retinas treated with 25 μm Rockout possessed significantly more pH3-positive Müller glia in the basal INL (Fig. 6A–I; 25.6 ± 8.5%, n = 18, p = 0.003) relative to DMSO controls (4.9 ± 1.8%, n = 27). These data suggest that inhibition of Rocks disrupted Müller glia INM in the light-damaged adult zebrafish retina.
Figure 6.
Inhibition of Rocks as well as rock2a and rock2b double knockdowns alter the position of pH3-positive Müller glia and of NPC nuclei after light damage. A–H, Single z-plane confocal images of albino;Tg[gfap:EGFP]nt11 zebrafish that were exposed to either DMSO (A, C, E, G) or 25 μm Rockout (B, D, F, H) were immunocytochemically labeled with pH3 (A, B) and PCNA (C, D). Inhibition of Rocks altered the position of pH3-positive Müller glia (A, B, G, H), but not the number of PCNA-positive cells (C, D, G, H) at 35 h after starting the light treatment. Histograms depict the distribution of pH3-positive nuclei in the ONL and apical and basal INL in DMSO- and Rockout-treated zebrafish at 35 h (I) and 45 h (J) after starting the light treatment. Data are shown as mean ± SE, n ≥ 18, Student's t test, *p < 0.05 between corresponding retinal regions in DMSO- and Rockout-treated samples. K, Immunoblot of phosphorylated MLC (pMLC, Thr18/Ser19) and actin as a loading control in DMSO and Rockout-treated retinal lysates. n = 3. L, RT-PCR was performed on cDNA obtained from embryos to determine the efficacy of the rock2a [top, 24 h postfertilization (hpf)] and rock2b (bottom, 72 hpf) splice junction morpholinos, respectively. M, N, Histograms of the distribution of pH3-positive nuclei in the retina depicting that significantly more pH3-positive nuclei were located in the basal INL in rock2a and rock2b double morphants relative to standard control morphants at 45 h (M) and 72 h (N) of light treatment. O–T, Single z-plane confocal images of albino;Tg[gfap:EGFP]nt11 zebrafish that were electroporated with either a standard control morpholino (O, Q, S; SC MO) or a combination of rock2a and rock2b splice site morpholinos (P, R, T), light-damaged for 72 h, and immunocytochemically labeled for pH3 (O, P, S, T) and GFP (Q–T) and counterstained with the nuclear dye DAPI (S, T). GCL, Ganglion cell layer; SC MO, standard control morpholino; wt, wild-type. Scale bar, 20 μm for A–H and O–T.
At time points when NPCs divide, pH3-positve nuclei predominantly localized to the ONL (Fig. 1Z). To determine whether NPCs also require Rock activity to migrate to the ONL, light-damaged zebrafish were exposed to either DMSO or 25 μm Rockout from 28 to 45 h after the start of the light treatment. Similar to Müller glia, the number of pH3-positive NPCs was significantly increased in the basal INL in the presence of 25 μm Rockout at 45 h of light treatment compared with DMSO controls (Fig. 6J; Rockout: 18.6 ± 4.9%, n = 22; DMSO: 2.8 ± 1.2%, n = 26, p = 0.0008). Accordingly, significantly fewer pH3-positive NPCs were observed in the ONL in Rockout-exposed retinas (Fig. 6J; 39.8 ± 7.4%, n = 22, p = 0.0007) relative to DMSO controls (73.8 ± 6.8%, n = 26). Similar changes in the distribution of pH3-positive cells were observed when retinas were exposed to 25 μm Rockout from 28 to 72 h (data not shown). Together, our data suggest that INM of both Müller glia and NPCs was facilitated by Rocks.
Phosphorylation of MLC is required for the function of myosin and consequently actin-myosin-mediated contraction (Vicente-Manzanares et al., 2009). Because MLC is one direct target of Rocks (Amano et al., 1996; Vicente-Manzanares et al., 2009), we performed immunoblot analysis to test whether exposure to Rockout inhibits MLC phosphorylation. Although exposure of retinas to Rockout resulted in reduced phosphorylated MLC levels compared with DMSO controls (Fig. 6K; 0.57 ± 0.05 and 1.0 ± 0.20, respectively, n = 3, p = 0.052), this reduction failed to reach statistical significance. Together, these data suggest that the disruption of actin-myosin-mediated contraction might be the underlying cause for the change in the distribution of pH3-positive nuclei along the apicobasal axis in Rockout-treated retinas.
Rock2a was previously implicated in mediating INM in the developing medaka retina (Herder et al., 2013). In the adult regenerating retina, we observed that rock2a and rock2b mRNAs were upregulated, although not significantly, at 16 and 35 h of light treatment. To confirm the Rockout results suggesting a role for Rocks in INM, we used splice site targeting morpholinos to knockdown the expression of Rock2a and Rock2b. Initially, RT-PCR analysis of mRNA from morpholino-injected embryos and light-damaged retinas was used to verify the efficacy of the morpholinos. Standard control morphants amplified the expected 653 bp product from mRNA that was obtained from either injected embryos at 24 h post fertilization or 35 h light-damaged retinas (Fig. 6L, top, and data not shown). In contrast, the rock2a morphant cDNA predominantly amplified a slightly shorter fragment (Fig. 6L, top, and data not shown). Sequence analysis of this shorter PCR product revealed that the rock2a morpholino caused the exclusion of exon 3 in the morphant mRNA, resulting in a premature translation stop codon and a protein with a disrupted catalytic domain (data not shown). Similarly, the rock2b splice site morpholino (Wang et al., 2011) caused a deletion of exon 3, resulting in a predominant PCR fragment that was shorter relative to the wild-type product observed in standard control morphants (Fig. 6L, bottom, expected PCR product size 550 bp, and data not shown). Deletion of exon 3 of rock2b morphant mRNA resulted in a premature translation stop codon that disrupted the catalytic domain of Rock2b. Having established that both rock2a and rock2b morpholinos successfully disrupted splicing, we examined next whether disrupting both rock2a and rock2b splicing altered the position of pH3-positive nuclei.
The rock2a and rock2b splice site targeting morpholinos were combined, intravitreally injected, and electroporated into albino;Tg[gfap:EGFP]nt11 zebrafish before they were exposed to constant intense light for 45 or 72 h. Similar to Rockout-exposed retinas, rock2 double morphant retinas contained significantly more pH3-positive nuclei in the basal INL relative to standard control morphants at both 45 and 72 h of light treatment (Fig. 6M–T; 45 h: rock2a+b: 20.8 ± 12.5%, n = 4; standard control: 0 ± 0%, n = 8, p = 0.016; 72 h: rock2a+b: 14.7 ± 6.9%, n = 14; standard control: 0 ± 0%, n = 11, p = 0.036). In contrast, the percentage of pH3-positive nuclei in the apical INL and the ONL were not significantly different between rock2 double morphant and standard control morphants at 45 (Fig. 6M; apical INL: rock2a+b: 41.7 ± 25.0%, n = 4; standard control: 31.3 ± 16.2%, n = 8, p = 0.36; ONL: rock2a+b: 37.5 ± 23.9%, n = 4; standard control: 68.8 ± 16.2%, n = 8, p = 0.15) and 72 h of light treatment (Fig. 6N–T; apical INL: rock2a+b: 12.3 ± 4.0%, n = 14; standard control: 13.7 ± 9.0%, n = 11, p = 0.44; ONL: rock2a+b: 72.0 ± 8.6%, n = 14; standard control: 86.3 ± 9.0%, n = 11, p = 0.15). The total number of pH3-positive cells was also not statistically different between the rock2 double morphant and standard control morphant retinas at both 45 and 72 h of light (45 h: rock2a+b: 0.60 ± 0.29 cells/300 μm, n = 13; standard control: 0.96 ± 0.30 cells/300 μm, n = 12, p = 0.40; 72 h: rock2a+b: 3.06 ± 0.61 cells/300 μm, n = 11; standard control: 4.43 ± 0.69 cells/300 μm, n = 15, p = 0.15). Together, these data suggest that Rock2a and Rock2b played roles in mediating INM in Müller glia and NPCs.
Inhibiting Rock2a and Rock2b reduces the number of proliferating cells
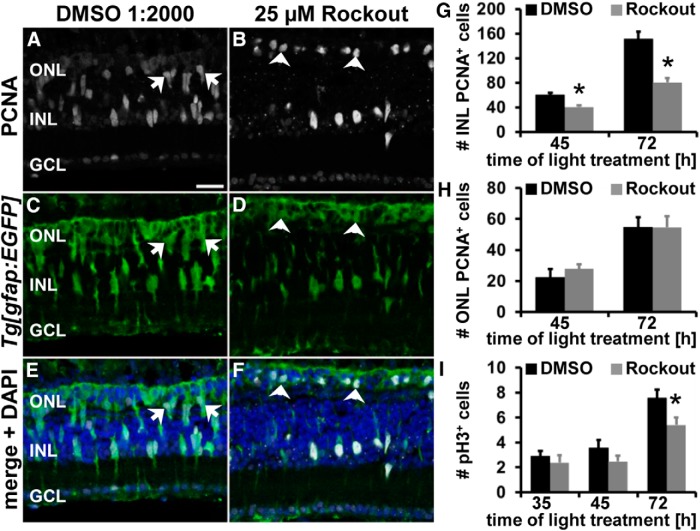
During retinal development, it was suggested that cells that do not divide apically exit the cell cycle prematurely (Del Bene et al., 2008). Therefore, we examined whether Rockout-mediated INM disruption affected Müller glia and NPC proliferation. Retinas exposed to Rockout from 28 to 35 h after starting the light treatment possessed similar numbers of PCNA-positive cells in both the INL and ONL (INL: 36.05 ± 1.86 cells/300 μm, n = 23, p = 0.91; ONL: 4.37 ± 0.82 cells/300 μm; n = 23, p = 0.055) relative to DMSO controls (INL: 36.4 ± 2.29 cells/300 μm, n = 31; ONL: 7.30 ± 1.23 cells/300 μm, n = 31). Although Rockout did not disrupt the induction of Müller glia proliferation, we examined the effect of Rockout on NPC proliferation. At 45 h of light treatment, significantly fewer PCNA-positive cells were present in the INL of Rockout-treated retinas (Fig. 7A–G; 40.74 ± 2.74 cells/300 μm, n = 13, p = 0.0026) relative to DMSO controls (60.65 ± 5.37 cells/300 μm, n = 16). This effect persisted at 72 h of constant intense light (Fig. 7G; Rockout: 80.55 ± 6.81 cells/300 μm, n = 20; DMSO: 151.81 ± 11.67 cells/300 μm, n = 18, p = 4.4 * 10−6). In contrast, the number of PCNA-positive cells in the ONL, which comprises both rod precursors and Müller glia/NPCs, were not significantly different in Rockout and DMSO-treated retinas at both 45 and 72 h of light treatment (Fig. 7H). Interestingly, many PCNA-positive Müller glia/NPC nuclei in the ONL of Rockout-exposed retinas had a round morphology at 45 h of constant intense light (arrowheads, Fig. 7B,D,F) relative to the elongated nuclei in DMSO-treated controls (arrows, Fig. 7A,C,E). Moreover, the gfap:EGFP-positive cells with nuclear PCNA in the ONL of Rockout-exposed retinas were smaller than the mitotic cells in DMSO controls. These data suggest that a subset of those gfap:EGFP-positive cells that migrated to the ONL to divide did not return to the INL, but rather remained in the ONL in Rock-inhibited retinas. To further assess this possibility, we quantified the number of gfap:EGFP cells with nuclear PCNA located in the ONL. Significantly more gfap:EGFP-positive cells containing nuclear PCNA were present in the ONL of Rockout-treated retinas relative to DMSO controls (Rockout: 21.16 ± 2.69 cells/300 μm, n = 13; DMSO: 11.70 ± 2.57 cells/300 μm, n = 12; p = 0.019). These data suggested that Rock inhibition affected basal migration in a subset of gfap:EGFP-positive cells.
Figure 7.
Rock inhibition reduces NPC proliferation without affecting the number of mitotic cells. A–F, Single z-plane confocal images of light-damaged albino;Tg[gfap:EGFP]nt11 zebrafish that were exposed to either DMSO (A, C, E) or 25 μm Rockout (B, D, F) and immunocytochemically labeled with PCNA (A, B) and EGFP (C–F) show reduced numbers of PCNA-positive cells at 45 h of light treatment. Arrows and arrowheads indicate PCNA-positive ONL nuclei with an elongated (A, C, E) or a round morphology (B, D, F), respectively. Histogram depicting the number of PCNA-positive cells in the INL (G) and ONL (H) in DMSO- and Rockout-treated retinas at 45 and 72 h of light treatment. I, Histogram displaying the number of pH3-positive cells in Rockout- and DMSO-exposed retinas at 35, 45, and 72 h after starting the light treatment. GCL, Ganglion cell layer. Data are shown as mean ± SE, n ≥ 12, Student's t test, *p < 0.05. Scale bar, 20 μm for A–F.
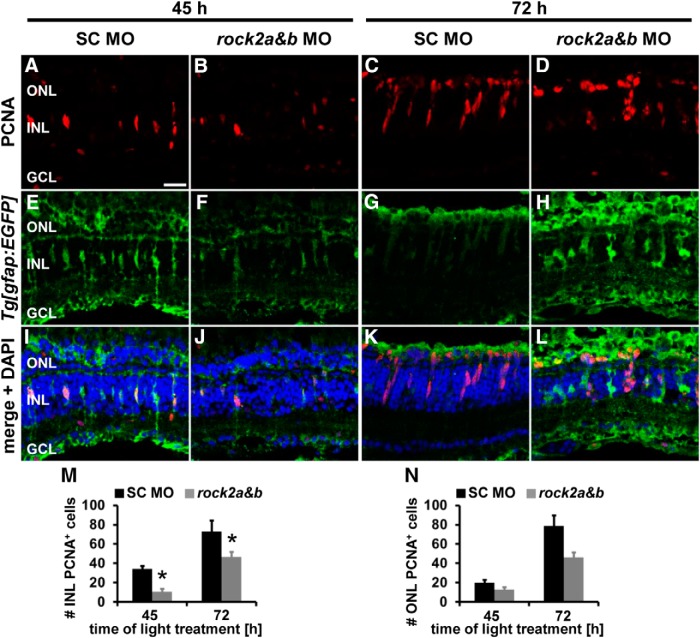
Having shown that rock2a and rock2b double knockdown caused a similar increased percentage of pH3-positive cells to remain in the basal INL as Rockout treatment, we also investigated whether the double knockdown affected the proliferative potential of the regenerating retina. At 45 h of light treatment, the number of PCNA-positive cells were significantly reduced in the INL, but not the ONL, of rock2 double morphants relative to standard control morphant retinas (Fig. 8A,B,I,J,M,N; INL: rock2a+b: 10.72 ± 2.69 cells/300 μm, n = 13; standard control: 34.32 ± 2.96 cells/300 μm, n = 11, p = 6.1 * 10−6; ONL: rock2a+b: 12.73 ± 2.29 cells/300 μm, n = 13; standard control: 19.87 ± 3.49 cells/300 μm, n = 11, p = 0.09). The majority of the PCNA-positive cells in both rock2 double morphant and standard control morphants were in a single-cell stage, suggesting that fewer Müller glia were induced to proliferate at 45 h of light treatment in the rock2 double morphant retinas. Significantly reduced numbers of PCNA-positive cells were also observed in the INL of rock2 double morphant compared with standard control morphant retinas at 72 h of light treatment (Fig. 8C,D,G,H,K,L,M; rock2a+b: 46.81 ± 4.86 cells/300 μm, n = 15; standard control: 72.90 ± 11.32 cells/300 μm, n = 10, p = 0.026), whereas similar numbers were present in the ONL (Fig. 8C,D,G,H,K,L,N; rock2a+b: 46.33 ± 10.86 cells/300 μm, n = 15; standard control: 78.45 ± 15.08 cells/300 μm, n = 10, p = 0.089). These data indicate that double knockdown of rock2a and rock2b induced fewer Müller glia to proliferate and delayed NPC proliferation in the light-damaged retina. Although this is similar to the Rock-inhibited retina, the greater numbers of proliferating cells in the Rockout-treated retina was likely due to the application of Rockout at 28 h after starting the light treatment relative to the morpholino injection/electroporation before the start of the light treatment. Therefore, the Rockout-treatment did not affect Müller glia induction and proliferation and permitted us to focus on the role of Rocks in NPCs.
Figure 8.
Delayed proliferation in rock2a and rock2b double morphants. A–L, Single z-plane confocal images of albino;Tg[gfap:EGFP]nt11 zebrafish that were either electroporated with a standard control morpholino (columns 1 and 3) or a combination of rock2a and rock2b splice site morpholinos (columns 2 and 4) before exposing them to constant intense light for either 45 (columns 1 and 2) or 72 h (columns 3 and 4). Retinal sections were immunocytochemically labeled for PCNA (A–D, I–L) and EGFP (E–L) and counterstained with DAPI (I–L). M, N, Histograms displaying the number of PCNA-positive cells in the INL (M) and ONL (N) at 45 and 72 h of light treatment, respectively, revealing that significantly fewer PCNA-positive cells were present in the INL but not in the ONL of rock2 double morphants relative to standard control morphants. Data are shown as mean ± SE, n ≥ 4, Student's t test, *p < 0.05 between corresponding SC MO and rock2a and rock2b MO. GCL, Ganglion cell layer. Scale bar, 20 μm for A–L.
Fewer NPCs undergo DNA replication in Rock-inhibited retinas
The observed reduction in INL proliferation after Rock inhibition could result from fewer cells progressing into mitosis, increased cell death, or premature cell cycle exit. We first tested whether Rock inhibition affected the transition from the S phase into mitosis by determining the total number of pH3-positive (mitotic) cells. Similar numbers of pH3-positive cells were present in Rockout- and DMSO-exposed retinas at 35 h of light treatment (Fig. 7I; 25 μm Rockout: 2.40 ± 0.59 cells/300 μm, n = 23; DMSO: 2.89 ± 0.42 cells/300 μm, n = 32, p = 0.49) and 45 h of light treatment (Fig. 7I; Rockout: 2.47 ± 0. 50 cells/300 μm, n = 17; DMSO: 3.58 ± 0.64 cells/300 μm, n = 16; p = 0.18). This suggested that Rockout did not significantly affect early mitotic events, primarily Müller glia and early NPC mitosis. In contrast, at 72 h of light treatment, the number of pH3-positive cells was significantly reduced in Rockout-treated retinas (Fig. 7I; 5.41 ± 0. 61 cells/300 μm, n = 20, p = 0.025) relative to DMSO controls (7.55 ± 0.69 cells/300 μm, n = 18), which likely reflects the significantly decreased numbers of proliferating NPCs at this time point. Inhibition of Rocks reduced proliferation without affecting the number of mitotic events.
Using the TUNEL assay, we investigated whether apoptosis of NPCs was the underlying cause for reduced proliferation in Rock-inhibited retinas. TUNEL was performed on retinal sections exposed to Rockout or DMSO from 28–55 or 72 h of light treatment. A similarly small number of TUNEL- and PCNA-double positive cells were observed in the INL and ONL of both Rockout- and DMSO-treated retinas at either 55 h (data not shown) or 72 h of light treatment (INL: Rockout: 0.88 ± 0.28 cells/300 μm, n = 24; DMSO: 0.43 ± 0.21 cells/300 μm, n = 20, p = 0.22; ONL: Rockout: 1.10 ± 0.43 cells/300 μm, n = 24; DMSO: 0.46 ± 0.24 cells/300 μm, n = 20, p = 0.23). Therefore, cell death was not the underlying cause of reduced proliferation in Rockout-exposed retinas.
During retinal development, disrupting INM causes nonapical cell divisions that results in premature cell cycle exit and depletion of the NPC pool (Del Bene et al., 2008). Therefore, NPCs commit to lineages that produce earlier born neurons, such as ganglion cells, at the expense of those born later (Del Bene et al., 2008). Therefore, we set out to determine whether inhibition of Rocks causes premature cell cycle exit using an EdU/BrdU pulse chase approach. Zebrafish were injected intraperitoneally with EdU before Rockout or DMSO exposure at 28 h of light treatment to label Müller glia that were induced to enter the cell cycle (Fig. 9A–C). At 44 h of light treatment, the EdU-treated zebrafish were intraperitoneally injected with BrdU to label the DNA for 1 h until 45 h of light treatment to identify the Müller-glia-derived NPCs that remained in the cell cycle (Fig. 9A,D,E). The Rockout-exposed light-damaged zebrafish possessed significantly fewer EdU- and BrdU-colabeled cells than DMSO controls (Fig. 9H; Rockout: 7.79 ± 1.64 cells/300 μm, n = 17; DMSO: 24.05 ± 3.61 cell/300 μm, n = 10, p = 8.6 * 10−5). Therefore, Rockout reduced the number of cells that actively underwent DNA replication during the 1 h of BrdU exposure. This significant difference persisted when EdU-labeled zebrafish were allowed to incorporate BrdU for an extended period from 44–50 h of light treatment (Fig. 9H; Rockout: 10.09 ± 1.77 cells/300 μm, n = 13; DMSO: 35.01 ± 6.13 cells/300 μm, n = 5, p = 6.3 * 10−5). Together, these data demonstrate that fewer cells underwent the S phase after inhibition of Rocks, indicating that cells might have exited the cell cycle prematurely either due to defective INM or loss of Rho-mediated signaling, preventing cell cycle re-entry. Although reduced proliferation observed in rock2a and rock2b double morphants would be consistent with the latter explanation, it is unlikely the cause because we did not observe a difference in the number of BrdU-only-positive cells in Rockout- and DMSO-treated retinas in the EdU/BrdU pulse chase experiments at either 45 h (Fig. 9I; Rockout: 6.06 ± 1.10 cells/300 μm, n = 17; DMSO: 6.66 ± 1.92 cells/300 μm, n = 10, p = 0.77) or 50 h of light treatment (Fig. 9I; Rockout: 11.42 ± 2.54 cells/300 μm, n = 13; DMSO: 6.31 ± 3.22 cells/300 μm, n = 5, p = 0.28). This population of BrdU-only-positive cells represents a group of Müller glia that was recruited into the cell cycle after EdU was cleared from the zebrafish (Nelson et al., 2012) during the period of Rock inhibition. Therefore, inhibiting Rock-mediated signaling did not affect the number of cells recruited into the cell cycle.
Figure 9.
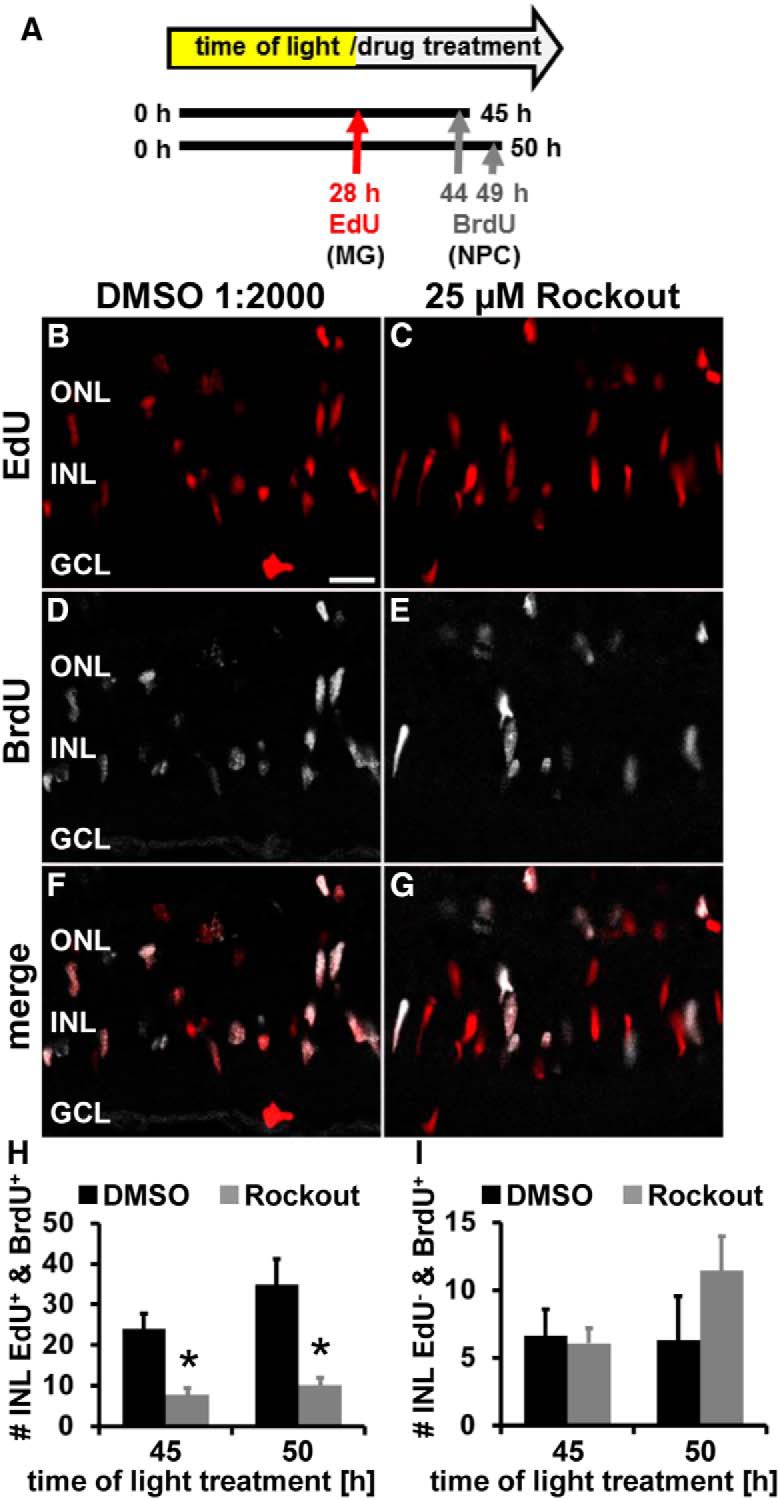
Rockout reduces the number of NPCs undergoing S-phase. A, Schematic of experimental design indicating that EdU was intraperitoneally injected at 28 h to label initially proliferating Müller glia and BrdU at 44 and 49 h of light treatment to label NPCs. Zebrafish were systemically exposed to DMSO or 25 μm Rockout starting at 28 h. B–G, Fewer EdU and BrdU double-positive cells were observed in single z-plane confocal images from light-damaged zebrafish exposed to 25 μm Rockout (C, E, G) relative to DMSO controls (B, D, F). Histogram depicting the number of EdU and BrdU double-positive INL cells (H) and the number of cells that are only BrdU-positive (I) in DMSO- and Rockout-treated retinas at 45 and 50 h after starting the light treatment. GCL, Ganglion cell layer. Data are shown as mean ± SE, n ≥ 6, Student's t test, *p < 0.05. Scale bar, 20 μm for B–G.
We further investigated the possibility that Rho-mediated signaling directly regulated cell cycle re-entry. The cell permeable Rho activator CN03 (5 μg/ml) or H2O was injected intravitreally into healthy, undamaged albino;Tg[gfap:EGFP]nt11 zebrafish eyes to test whether activation of Rho-signaling was sufficient to induce proliferation. At 16 h after injection, CN03 treatment did not increase proliferation in either the INL or ONL in healthy albino;Tg[gfap:EGFP]nt11 zebrafish retinas relative to H2O controls (Fig. 10A,B,I). These data suggest that Rho activation was not sufficient to induce Müller glia proliferation. Therefore, Rocks unlikely regulated the decision to remain in or exit the cell cycle directly, but rather mediated INM that resulted in early cell cycle exit when disrupted.
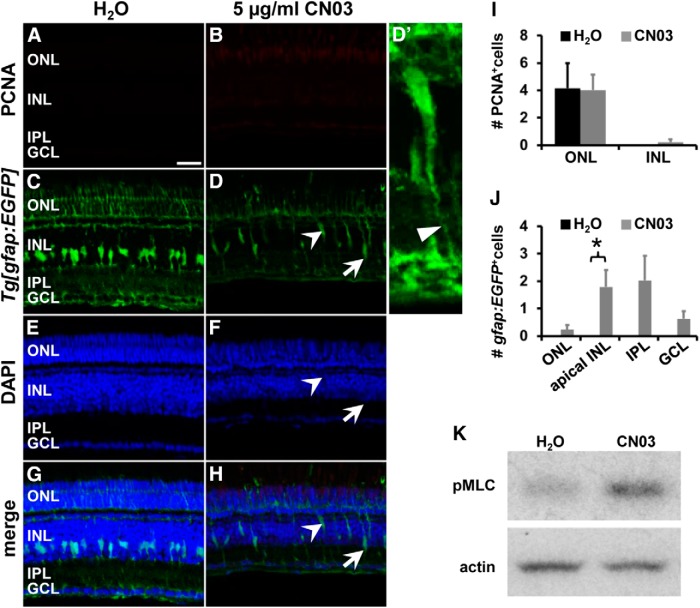
Figure 10.
Rho activation by CN03 stimulates mislocalization of Müller glia but does not induce proliferation at 16 h. A–H, Single z-plane confocal images of undamaged albino;Tg[gfap:EGFP]nt11 zebrafish that were intravitreally injected with either H2O (A, C, E, G) or 5 μg/ml CN03 (B, D, F, H) were immunocytochemically labeled for PCNA (A, B) and EGFP (C, D) and revealed that Rho activation caused Müller glia to migrate both apically within the INL (arrowhead, D, F, H) and basally into the IPL (arrow, D, F, H) at 16 h after exposure. (D′) Magnified 3D projection of the apically migrated cell indicated by the arrowhead in (D, F, H) showing that the cell maintains its basal process (thin arrowhead). CN03 did not induce an upregulation of PCNA-positive cells after 16 h of treatment in either the ONL or INL in undamaged retinas (I). Histogram depicting the number of gfap:EGFP-positive cells in the ONL, apical INL, IPL, and ganglion cell layer (GCL) (J). Data are shown as mean ± SE, n ≥ 11, Student's t test, *p < 0.05 between corresponding retinal regions in H2O- and CN03-treated samples. K, Immunoblot of phosphorylated MLC (pMLC, Thr18/Ser19) and actin as a loading control in H2O- and CN03-treated retinal lysates after 16 h. Scale bar, 20 μm for A–H.
In support of Rho signaling playing a role in INM, at 16 h after intravitreal injection of 5 μg/ml CN03 into undamaged eyes, a small but significantly increased number of Müller glia soma were mislocalized to the apical INL relative to H2O controls (Fig. 10C–H,J; CN03: 1.79 ± 0.61 cells/300 μm, n = 11; H2O: 0 ± 0 cells/300 μm, n = 8, p = 0.025). In addition, a few cells had extended into the IPL in CN03-treated retinas (Fig. 10D,H,J). Although a redistribution of gfap:EGFP-positive cells within the retina was observed after Rho activation by CN03, the total number of gfap:EGFP-positive Müller glia was similar to H2O-injected retinas (Fig. 10C–H; data not shown). Müller glia that mislocalized to the apical INL maintained their basal processes (Fig. 10D′), suggesting that detachment from the basement membrane was not the underlying cause for their apical displacement. In addition, we observed that rod photoreceptors migrated apically interspersing with cone photoreceptors, whereas other cell types such as HuC/D-positive amacrine/ganglion cells and PKCα-positive bipolar cells did not change their relative position (data not shown). To test whether this migratory phenotype might be due to Rho-mediated phosphorylation of MLC, we determined the levels of phosphorylated MLC by immunoblot. Intravitreal injections of CN03 induced higher levels of phosphorylated MLC relative to H2O-injected controls (Fig. 10K), indicating that the change in Müller glial position might have resulted from increased MLC phosphorylation. Together, these data support a role for Rho-mediated signaling events in regulating the migration of gfap:EGFP-positive Müller glia soma, likely through the upregulation of phosphorylated/activated MLC similar to that observed during INM in the light-damaged retina.
Inhibition of Notch signaling does not rescue NPC proliferation
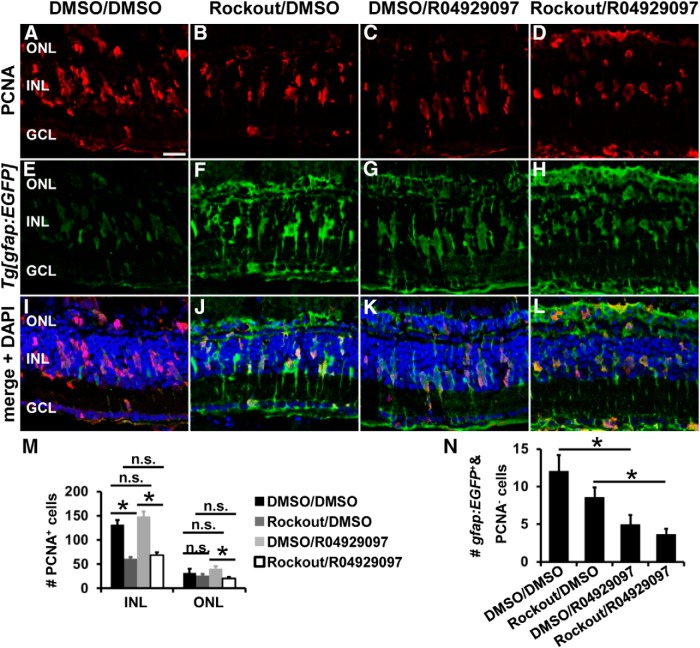
Disrupting INM in the developing retina resulted in premature cell cycle exit of NPCs, which was suggested to occur due to reduced exposure to Notch signaling because overexpression of the Notch intracellular domain promoted NPCs to remain in the cell cycle (Del Bene et al., 2008). In the regenerating adult retina, however, inactivation of Notch signaling is required to recruit Müller glia into the cell cycle (Wan et al., 2012; Conner et al., 2014). Therefore, we investigated whether the arising NPCs in the Rock-inhibited retina can be retained in the cell cycle by inhibiting Notch signaling using the γ-secretase inhibitor R04929097. Light-damaged albino;Tg[gfap:EGFP]nt11 zebrafish were intraperitoneally injected with the γ-secretase inhibitor R04929097 (750 μm) or the vehicle control DMSO (10%) before exposing them systemically to either 25 μm Rockout or DMSO (1:2000) starting at 28 h of light treatment. Retinas of the Rockout-exposed treatment groups (Rockout/DMSO and Rockout/R04929097) contained significantly fewer PCNA-positive cells relative to their corresponding DMSO treatment groups (DMSO/DMSO, DMSO/R04929097) at 72 h of light treatment (Fig. 11A–M). However, inhibition of Notch activation by R04929097 was not sufficient to induce increased numbers of PCNA-positive cells in Rockout-exposed retinas relative to those treated with Rockout/DMSO at 72 h of light treatment (Fig. 11B,D,J,L,M; Rockout/R04929097: 68.4 ± 5.7, n = 16; Rockout/DMSO: 61.1 ± 3.2, n = 23; Tukey's test, p = 0.87). Surprisingly, R04929097 treatment of DMSO-exposed zebrafish also did not increase the number of proliferating cells compared with the corresponding DMSO control (Fig. 11A,C,I,K,M; DMSO/R04929097: 148.1 ± 10.9, n = 18; DMSO/DMSO: 131.9 ± 9.2, n = 13; Tukey's test, p = 0.48). The number of PCNA-positive cells in the ONL was not statistically significant between the DMSO/DMSO-treated zebrafish and the other treatment groups (Fig. 11M; Rockout/R04929097: 20.1 ± 3.6, n = 16, Tukey's test: pDMSO/DMSO = 0.21, pRockout/DMSO = 0.55; Rockout/DMSO: 26.7 ± 3.1, n = 23, Tukey's test: pDMSO/DMSO = 0.78; DMSO/R04929097: 40.8 ± 4.6, n = 18, Tukey's test: pDMSO/DMSO = 0.36; DMSO/DMSO: 31.5 ± 9.2, n = 13). To ensure that the R04929097 treatment paradigm was successful, we determined the number of Müller glia that were not in the cell cycle (gfap:EGFP-positive and PCNA-negative cells) at 72 h of light treatment. R04929097 significantly reduced the numbers of gfap:EGFP-positive PCNA-negative cells compared with DMSO in both Rockout and DMSO-treated retinas (Fig. 11E–L,N; Rockout/R04929097: 3.7 ± 0.7, n = 16, Tukey's test: pDMSO/DMSO = 0.001, pRockout/DMSO = 0.046; DMSO/R04929097: 5.0 ± 1.2, n = 18, Tukey's test: pDMSO/DMSO = 0.004; Rockout/DMSO: 8.6 ± 1.3, n = 23, Tukey's test: pDMSO/DMSO = 0.29; DMSO/DMSO: 12.1 ± 2.1, n = 13), indicating that R04929097 treatment significantly increased the number of Müller glia in the cell cycle. Together, inhibition of Notch signaling by R04929097 induced more Müller glia to re-enter the cell cycle in both Rockout- and DMSO-exposed retinas, but did not rescue proliferation of NPCs in the Rock-inhibited retina.
Figure 11.
Notch inhibition by R04929097 does not increase NPC proliferation in Rockout-treated retinas at 72 h of light treatment. A–L, Single z-plane confocal images of light-damaged albino;Tg[gfap:EGFP]nt11 zebrafish that were intraperitoneally injected either with R04929097 (columns 3 and 4) or DMSO (columns 1 and 2) before exposing them to either Rockout (column 2 and 4) or DMSO (columns 1 and 3) at 28 h of light treatment. Retinas collected at 72 h of light treatment were immunocytochemically labeled for PCNA (A–D, I–L) and GFP (E–L) and counterstained with DAPI (I–L). M, Histogram depicting the number of PCNA-positive cells in the INL and ONL at 72 h of light treatment, revealing that treating Rockout-exposed zebrafish with the γ-secretase inhibitor R04929097 did not increase the number of proliferating cells significantly. N, Histogram revealing that the number of gfap:EGFP-positive cells that were PCNA-negative was significantly decreased in the R04929097-treated DMSO- or Rockout-exposed zebrafish compared with those that received DMSO, which indicated that more Müller glia were recruited into the cell cycle in response to R04929097. Data are shown as mean ± SE, n ≥ 13, ANOVA followed by Tukey's post hoc test, *p < 0.05 as indicated by bars spanning between the treatment groups (M, N). n.s., Not significant; GCL, ganglion cell layer. Scale bar, 20 μm for A–L.
Rock inhibition reduces neuronal commitment of ONL proliferating cells
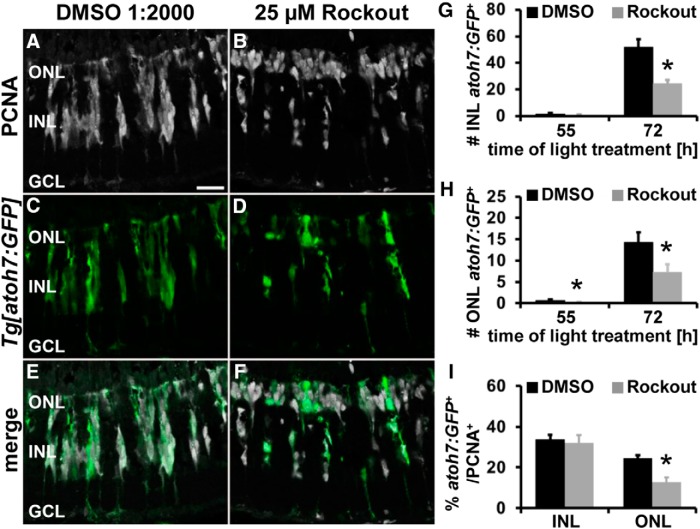
During retinal development, defective INM causes early cell cycle exit and increased expression of neuronal commitment factors such as atoh7 (Del Bene et al., 2008). Therefore, we investigated whether the cells that exited the cell cycle in Rockout-exposed retinas committed prematurely to a neuronal lineage in the light-damaged retina. It was shown that cells committing to the photoreceptor lineage express the transcription factor atoh7 both during retinal development (Poggi et al., 2005) and regeneration of light-damaged retinas (Conner et al., 2014). In the regenerating retina, atoh7:GFP expression is initially observed in single PCNA-positive cells at 51 h of light treatment (M.L. and D.R.H., unpublished observation). Therefore, to determine whether Rockout-dependent suppression of INM induced NPCs to commit earlier to the photoreceptor lineage, albino;Tg[atoh7:GFP]rw021 zebrafish were treated with either 25 μm Rockout or 1:2000 DMSO from 28–45 h of light treatment. At 45 h, we occasionally observed an atoh7:GFP-positive cell in both 25 μm Rockout- and DMSO-exposed retinas (data not shown). Therefore, Rockout treatment did not cause NPCs to commit prematurely to the neuronal lineage at 45 h of light treatment. Because the phenotype might only become evident at a later time, we investigated the commitment of NPCs to the neuronal lineage after exposure to Rockout from 28 to 55 h of light treatment. At 55 h of light damage, similar numbers of single atoh7:GFP-positive cells were present in PCNA-positive clusters in Rockout-treated retinas in the INL relative to controls (Fig. 12G; Rockout: 0.69 ± 0.42/300 μm, n = 12; DMSO: 1.79 ± 0.54/300 μm, n = 11, p = 0.12). In contrast, a significant reduction in the number of atoh7:GFP-positive cells was observed in the ONL after Rockout exposure compared with DMSO controls (Fig. 12H; Rockout: 0.13 ± 0.09 cells/300 μm, n = 12; DMSO: 0.63 ± 0.20 cells/300 μm, n = 11; p = 0.03). These data indicate that proliferating cells did not commit earlier to the photoreceptor lineage. However, because the number of atoh7:GFP-positive cells was low at 55 h, we further examined their presence at 72 h. At this time point, significantly fewer atoh7:GFP-positive cells were present in the ONL in Rockout-exposed retinas (Fig. 12D,F,H; 7.33 ± 1.81 cells/300 μm, n = 20, p = 0.025) relative to DMSO controls (Fig. 12C,E,H; 14.22 ± 2.38 cells/300 μm, n = 18). A similar significant reduction in the number of atoh7:GFP-positive cells was observed in the INL of Rockout-exposed retinas (Fig. 12D,F,G; 24.56 ± 2.62 cells/300 μm, n = 20, p = 6.3 * 10−5) relative to DMSO controls (Fig. 12C,E,G; 52.01 ± 5.72 cells/300 μm, n = 18). The decreased number of atoh7:GFP-positive cells in Rockout-treated retinas could be a consequence of the reduced number of proliferating cells after inhibition of Rocks. Indeed, presenting the atoh7:GFP-positive cells as the percentage of PCNA-positive cells revealed that there was no significant difference in the INL between Rockout- and DMSO-treated retinas at 72 h of light damage (Fig. 12I; Rockout: 32.13 ± 3.86%/300 μm, n = 20; DMSO: 33.81 ± 2.17%/300 μm, n = 18, p = 0.72). In contrast, the number of atoh7:GFP-positive cells expressed as the percentage of PCNA-positive cells in the ONL was significantly lower in Rockout-exposed retinas (Fig. 12I; 12.66 ± 2.37%/300 μm, n = 20, p = 0.0003) relative to DMSO controls (24.52 ± 1.54%/300 μm, n = 18). These data show that although a higher degree of cell cycle exit was observed in Rock-inhibited retinas, the INL-based NPCs committed to the neuronal lineage in a similar time frame as controls. In contrast, Rock inhibition disrupted lineage commitment of NPCs in the ONL, potentially due to exposure or lack of regulating factors.
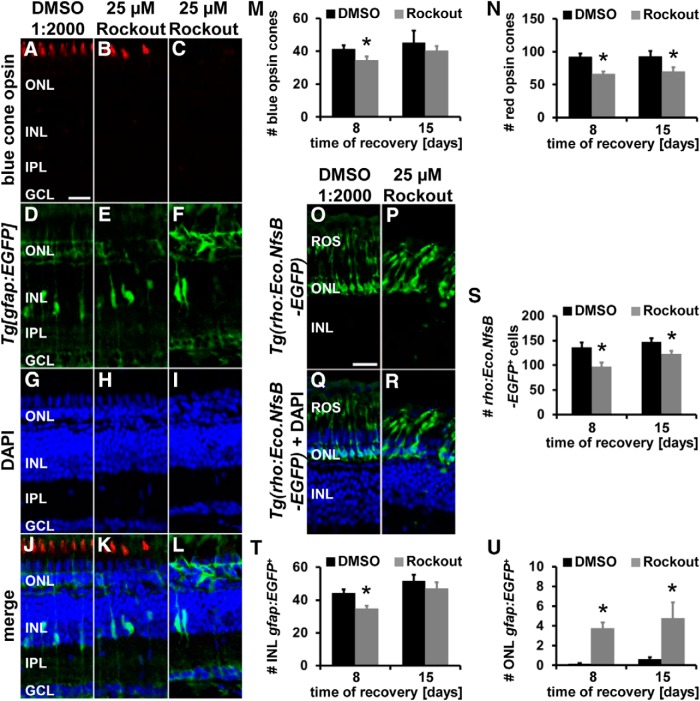
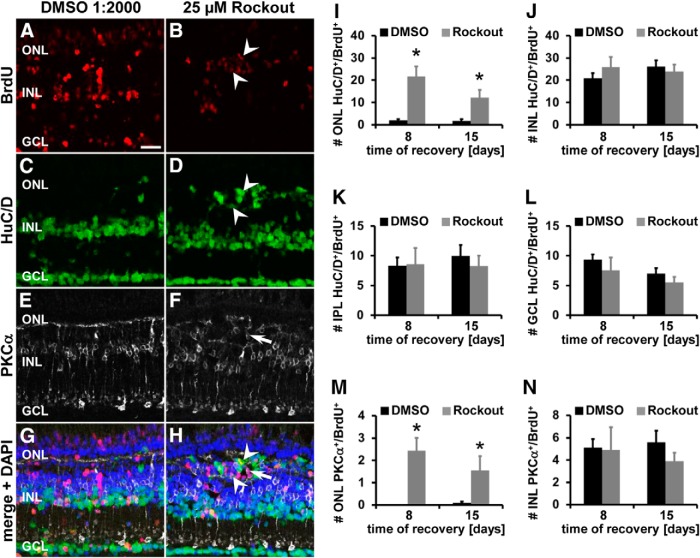
Figure 12.
Rock inhibition affects neuronal lineage commitment of ONL, but not INL, proliferative cells. A–F, Single z-plane confocal images of light-damaged albino;Tg[atoh7:GFP]rw021 zebrafish that were systemically exposed to either DMSO (A, C, E) or 25 μm Rockout (B, D, F) and immunocytochemically labeled for PCNA (A, B, E, F) and EGFP (C–F) show fewer atoh7:GFP-positive cells in Rockout-treated retinas relative to DMSO controls at 72 h of light treatment. Histogram depicting the number of atoh7:GFP-positive cells in the INL (G) and ONL (H) at 55 and 72 h of light treatment. I, Histogram displaying that the percentage of PCNA-positive cells that express atoh7:GFP in the ONL was significantly reduced, but not in the INL, of Rockout-treated retinas compared with DMSO controls at 72 h of light treatment. GCL, Ganglion cell layer. Data are shown as mean ± SE, n ≥ 18, Student's t test,*p < 0.05. Scale bar, 20 μm for A–F.
Rock inhibition affects regeneration of both cone and rod photoreceptors
Having observed reduced numbers of proliferating cells after disruption of INM by Rockout, we investigated the regeneration of cone photoreceptors after inhibiting Rocks from 28–72 h after starting the light treatment. Light-damaged albino;Tg[gfap:EGFP]nt11 zebrafish were exposed to 25 μm Rockout (corresponding to 28–72 h of light treatment) or its vehicle control DMSO (1:2000) and allowed to recover in system water for 8 or 15 d. Immunocytochemical labeling revealed that the number of both red- and blue-opsin-expressing cones were significantly reduced in Rockout-treated retinas after 8 d of recovery (Fig. 13A–C,M,N; blue: 34.39 ± 2.47 cells/300 μm, n = 25, p = 0.041; red: 65.81 ± 3.89 cells/300 μm, n = 19, p = 0.0003) relative to DMSO controls (Fig. 13M,N; blue: 41.29 ± 2.21 cells/300 μm, n = 29; red: 92.0 ± 5.27 cells/300 μm, n = 19). After 15 d of recovery, this effect persisted for red opsin cones (Fig. 13N; Rockout: 69.75 ± 6.52 cells/300 μm, n = 17; DMSO: 92.78 ± 8.17 cells/300 μm, n = 13, p = 0.034), but blue cone numbers were nearly the same in Rockout- and DMSO-treated retinas (Fig. 13M; Rockout: 40.29 ± 2.73 cells/300 μm, n = 12; DMSO: 45.18 ± 7.28 cells/300 μm, n = 11, p = 0.52). In addition, we examined the regeneration of rod photoreceptors in albino;Tg[rho:Eco.nfsB-EGFP]nt19 zebrafish (Montgomery et al., 2010) using the same paradigm. The number of EGFP-positive rod photoreceptors was significantly decreased in Rockout-exposed retinas relative to DMSO controls at 8 d of recovery (Fig. 13O–S; Rockout: 97.36 ± 7.99 cells/300 μm, n = 11; DMSO: 136.29 ± 10.25 cells/300 μm, n = 13, p = 0.008) and 15 d of recovery (Fig. 13S; Rockout: 123.21 ± 6.48 cells/300 μm, n = 15; DMSO: 147.17 ± 7.32 cells/300 μm, n = 13, p = 0.021). To determine whether Rockout exposure caused increased ONL cell death that led to the observed reduction in the number of photoreceptors at 8 and 15 d of recovery, retinal sections at 55 and 72 h of light treatment were subjected to the TUNEL assay. The numbers of TUNEL-positive cells were similar in the ONL of Rockout and DMSO-treated retinas at 55 h after starting the light treatment (Rockout: 29.40 ± 5.71 cells/300 μm, n = 10; DMSO: 17.59 ± 4.79 cells/300 μm, n = 10, p = 0.13). However, 72 h after starting the light treatment, significantly more TUNEL-positive cells were observed in the ONL in Rockout-exposed retinas relative to DMSO controls (Rockout: 18.43 ± 2.54 cells/300 μm, n = 24; DMSO: 8.68 ± 2.44 cells/300 μm, n = 20, p = 0.009). Together, these data suggest that both decreased proliferation resulting from Rock inhibition and increased ONL cell death reduced the regeneration of both cone and rod photoreceptors.
Figure 13.
Exposure to Rockout affects cone and rod photoreceptor regeneration and the localization of gfap:EGFP-positive Müller glia. A–L, O–R, Single z-plane confocal images of light-damaged albino;Tg[gfap:EGFP]nt11 (A–L) or albino;Tg[rho:Eco.Nfsb-EGFP]nt19 zebrafish (O–R) that were systemically exposed to either DMSO (A, D, G, J, O, Q) or 25 μm Rockout (B, C, E, F, H, I, K, L, P, R) from 28–72 h of light treatment, allowed to recover for 8 d, and immunocytochemically labeled with an antibody to blue cone opsin (A–C, J–L) and EGFP (D–F, J–L, O–R). Fewer blue cone (A–C, J–L) and rod photoreceptors (O–R) are regenerated and a subset of Müller glia is mislocalized to the ONL (D–F, J–L) at 8 d of recovery in Rockout-treated retinas relative to DMSO controls. Images of two different Rockout-treated retinas are presented in B, C, E, F, H, I, K, and L to display the variability in retinal recovery after similar exposure to Rockout. Histograms depicting the number of blue cones (M), red cones (N), rho:Eco.Nfsb-EGFP-positive rod photoreceptors (S), and gfap-EGFP-positive Müller glia in the INL (T) and ONL (U) at 8 and 15 d of recovery after exposure to DMSO or Rockout from 28–72 h. GCL, Ganglion cell layer; ROS, rod outer segment. Data are shown as mean ± SE, n ≥ 11, Student's t test, *p < 0.05. Scale bar, 20 μm for A–L and O–R.
Mislocalization of Müller glia at 8 and 15 d of recovery after Rock inhibition during Müller glia/NPC proliferation
Because Müller glia dedifferentiated into NPCs to regenerate lost photoreceptors, we investigated whether the pool of Müller glia was replenished during retinal regeneration. At 8 and 15 d of recovery, gfap:EGFP-positive Müller glia were observed in their typical basal location in the INL and their processes extended to both the ONL and ganglion cell layer in DMSO-exposed retinas (Fig. 13D and data not shown). In contrast, gfap:EGFP-labeled Müller glial processes in Rockout-exposed retinas were often disorganized in the ONL, but appeared normal in the ganglion cell layer (Fig. 13E,F). Furthermore, gfap:EGFP-positive cells in the INL were unevenly spaced giving rise to larger gaps in Rockout-treated retinas compared with DMSO controls (Fig. 13D–F). Quantification confirmed that the number of gfap:EGFP-positive cells in the INL was significantly reduced in Rockout-exposed retinas (Fig. 13T; 34.82 ± 1.75 cells/300 μm, n = 25, p = 0.002) compared with DMSO controls (Fig. 13T; 44.32 ± 2.25 cells/300 μm, n = 28) at 8 d of recovery. However, there was no significant difference in the number of INL-based gfap:EGFP-positive cells at 15 d of recovery (Fig. 13T; Rockout: 47.08 ± 3.81 cells/300 μm, n = 12; DMSO: 51.68 ± 3.93 cells/300 μm, n = 11, p = 0.41). Surprisingly, at both 8 and 15 d of recovery, significantly more gfap:EGFP-positive cells were localized in the ONL in Rockout-exposed retinas (Fig. 13D–F,U; 8 d: 3.75 ± 0.60 cells/300 μm, n = 25, p = 5.9 * 10−8; 15 d: 4.77 ± 1.60 cells/300 μm, n = 12, p = 0.022) relative to DMSO controls (8 d: 0.16 ± 0.07 cells/300 μm, n = 28; 15 d: 0.61 ± 0.23 cells/300 μm, n = 11). However, the total number of gfap:EGFP-positive cells did not differ between Rockout- and DMSO-treated retinas at 8 and 15 d recovery (data not shown). Together, inhibition of Rocks between 28 and 72 h of light treatment affected the localization of Müller glia, but not their differentiation.
Rockout treatment results in mislocalization of a subset of newly generated amacrine/ganglion and bipolar cells
During retinal development, disrupting INM causes an increased generation of early born retinal neurons such as ganglion and amacrine cells (Del Bene et al., 2008). Therefore, we investigated whether reduced regeneration of rod and cone photoreceptors resulted from incorrect differentiation of proliferating NPCs into ganglion or amacrine cells in Rock-inhibited retinas. We demonstrated previously that inner retinal neurons are not significantly lost by light damage (Vihtelic et al., 2006). We injected BrdU at 28, 48, 58, 84, 108, and 120 h after starting the light treatment to identify newly generated ganglion and amacrine cells in DMSO- and Rockout-exposed retinas at 8 and 15 d of recovery. Retinal sections that were immunocytochemically labeled for the ganglion and amacrine cell marker HuC/D (Wei et al., 2006; Conner et al., 2014) revealed an accumulation of misplaced HuC/D-positive cells between the horizontal cell layer and the rod photoreceptor cell layer (subsequently referred to as ONL HuC/D-positive cells) in Rock-inhibited retinas, of which a subset colabeled with BrdU at 8 d of recovery (Fig. 14A–D,G,H). There were significantly more HuC/D- and BrdU-double-positive cells located in the ONL of Rock-inhibited retinas relative to DMSO controls at 8 d of recovery (Fig. 14I; Rockout: 21.6 ± 4.6 cells/300 μm, n = 12; DMSO: 1.9 ± 0.6 cells/300 μm, n = 16, p = 4.5 * 10−5) and 15 d of recovery (Fig. 14I; Rockout: 12.2 ± 3.4 cells/300 μm, n = 17; DMSO: 1.8 ± 0.8 cells/300 μm, n = 18, p = 0.005). However, similar numbers of HuC/D- and BrdU-double-positive cells were present in the INL, IPL, and ganglion cell layer of both Rockout- and DMSO-exposed retinas (Fig. 14J–L). We also investigated whether Rockout treatment altered bipolar cell generation at 8 and 15 d of recovery. We immunocytochemically labeled retinal sections from BrdU-pulsed zebrafish for the ON-bipolar cell marker PKCα (Haverkamp et al., 2003; Conner et al., 2014). Similar, to HuC/D-positive cells, we observed increased numbers of misplaced PKCα- and BrdU-double-positive cells in the ONL in Rockout-exposed retinas relative to DMSO controls at 8 d of recovery (Fig. 14E–H,M; Rockout: 2.4 ± 0.6 cells/300 μm, n = 12; DMSO: 0 ± 0 cells/300 μm, n = 16, p = 3.8 * 10−5) and 15 d of recovery (Fig. 14M; Rockout: 1.6 ± 0.6 cells/300 μm, n = 17; DMSO: 0.09 ± 0.06 cells/300 μm, n = 18, p = 0.03). In contrast, the number of newly generated PKCα-positive cells in the INL did not differ between Rockout- and DMSO-treated retinas at 8 and 15 d of recovery (Fig. 14N). The generation of misplaced ganglion/amacrine and bipolar cells could have resulted from increased cell death in Rock-inhibited retinas. However, similar numbers of TUNEL-positive INL cells were observed in both DMSO- and Rockout-treated retinas at 55 and 72 h of light treatment (data not shown). Inhibiting Rocks caused incorrect differentiation of proliferating cells into misplaced amacrine/ganglion and bipolar cells, potentially at the expense of photoreceptors.
Figure 14.
Increased generation of HuC/D- and PKCα-positive neurons in the basal ONL in Rock-inhibited retinas. A–H, Single z-plane confocal images of light-damaged albino zebrafish that were systemically exposed to either DMSO (A, C, E, G) or 25 μm Rockout (B, D, F, H) from 28–72 h of light treatment. Zebrafish intraperitoneally injected with BrdU were allowed to recover for 8 d and were immunocytochemically labeled for BrdU (A, B, G, H), HuC/D (C, D, G, H), and PKCα (E–H). Rock inhibition caused an accumulation of HuC/D- and PKCα-positive cells in the basal ONL region and a subset of these cells colabeled with BrdU (arrowheads and arrow, respectively). I–N, Histograms depicting the number of HuC/D and BrdU double-positive cells in the ONL (I), INL (J), IPL (K), and ganglion cell layer (GCL; L) and the number of PKCα and BrdU double-positive cells in the ONL (M) and INL (N) at 8 and 15 d of recovery in retinas exposed to DMSO or Rockout from 28–72 h of light treatment. Data are shown as mean ± SE, n ≥ 12, Student's t test,*p < 0.05. Scale bar, 20 μm for A–H.
Discussion
We demonstrate that the majority of Müller glia and NPCs undergo INM to divide in the ONL of the regenerating zebrafish retina. Furthermore, filamentous actin structures form at the onset of INM at the rear of those Müller glia undergoing INM and both the actin cytoskeleton and Rocks are critical in facilitating apical migration of Müller glia nuclei, likely through MLC phosphorylation. Rock function is also required for basal migration of Müller glia and NPC nuclei. Although Rock2a and Rock2b double knockdown induces fewer Müller glia to re-enter the cell cycle, Rockout treatment starting at 28 h of light does not affect Müller glia proliferation or their progression through mitosis. However, Rock inhibition reduces the number of proliferating NPCs due to early cell cycle exit that is not accompanied by premature induction of the neuronal commitment factor atoh7. Long-term, Rocks are required to regenerate specific retinal neurons that were lost during light damage and to reestablish the correct localization of Müller glia, potentially by mediating INM.
In light-damaged retinas, mitotic nuclei (pH3-positive) were present in the ONL and apical INL when Müller glia divided, whereas they were predominantly located in the ONL during NPC divisions. Live-cell imaging confirmed that the majority of the gfap:EGFP-positive NPC divisions occurred in the ONL. In contrast, it was previously suggested that NPCs divide nonapically in light-damaged retinas based on the observation of pH3-positive nuclei in the ONL only early in the regeneration time course (Nagashima et al., 2013). However, at 3 d after initiating the light treatment, clusters of PCNA-positive cells in the INL and the majority of pH3-positive cells in the ONL were observed in both our data and in Nagashima et al. (2013). This confirmed that the retinas in both light damage models were at similar points of regeneration and that NPCs underwent INM to divide in the ONL. Previously, NPCs were shown to upregulate the expression of several genes preferentially, such as the transcription factors pax6a, pax6b, olig2, and neuroD, relative to Müller glia (Bernardos et al., 2007; Thummel et al., 2008b; Thummel et al., 2010; Thomas et al., 2012). Therefore, Müller glia and NPCs might express genes differentially that facilitate INM in the NPCs. Alternatively, space restrictions due to the presence of dying photoreceptors in the ONL that had not cleared when Müller glia divide, could explain the increased rate of cell divisions in the apical INL compared with the ONL-based NPC mitosis. This is consistent with retinal development, in which the position of mitosis changes from apical to subapical due to overcrowding of the apical surface at the onset of ONL formation (Weber et al., 2014).
Live-cell imaging of light-damaged retinal explants also revealed that the cleavage plane of cytokinesis of the majority of cells was horizontal. This type of cell division also occurs during early retinal development, with the orientation shifting vertically in a subset of NPCs at later stages. This change in the mitotic division plane was correlated with cell fate choices (Cayouette and Raff, 2003; Weber et al., 2014). We also observed that ∼10% of cells divided vertically; however, it is currently unknown whether the orientation of cell division affects cell fate decisions in the adult regenerating retina.
In the regenerating adult zebrafish retina, apical migration of Müller glia is mediated by the actin cytoskeleton and Rocks, which both regulate INM during retinal development (Murciano et al., 2002; Norden et al., 2009; Herder et al., 2013). Because Rock inhibition reduced the levels of MLC phosphorylation, Rocks likely control INM through the phosphorylation/activation of MLC, which is required for actin-myosin-mediated contraction (Ikebe et al., 1988). Different Rock target proteins that regulate the reorganization of the actin cytoskeleton could work in concert with MLC phosphorylation to facilitate INM (Nakai et al., 1997; Matsui et al., 1998; Maekawa et al., 1999). Because inhibiting either actin filament formation or Rocks, as well as rock2 morpholino knockdowns, did not completely block INM, either other signaling pathways facilitate actin/myosin contraction or different cytoskeletal components, such as microtubules, mediate apical INM (Del Bene et al., 2008; Yu et al., 2011). For example, apical nuclear migration in the developing chick neural tube requires both the actin cytoskeleton and microtubules (Spear and Erickson, 2012).
A subset of Müller glia/NPCs accumulated in the ONL after Rock inhibition. Similarly, exposing the developing telencephalon to low concentrations of the nonmuscle myosin inhibitor blebbistatin caused defects in NPC apical to basal migration (Schenk et al., 2009). These data suggest that both apical and basal migrations are actively driven processes. The velocity of basal INM would support that this process is actively driven in contrast to neuroepithelial development, where basal INM is thought to occur passively based on the velocity and mean squared displacement (Baye and Link, 2007; Kosodo et al., 2011; Leung et al., 2011). Rather than affecting nuclear migration directly, Rock/myosin may function to establish the basal process in daughter cells that lack it. Therefore, NPCs lacking basal processes cannot translocate their nuclei basally, leading to their accumulation in the ONL/apical surface. A similar phenotype was described in the developing mouse brain after knockdown of contactin due to failure to maintain/establish basal processes (Okamoto et al., 2013).
Reduced numbers of proliferating cells were observed in rock2 double morphant and Rock-inhibited retinas relative to control retinas. The rock2 double morphants likely possessed fewer Müller glia that re-entered the cell cycle due to the knockdowns beginning before starting the light treatment, whereas Rock inhibition caused reduced numbers of proliferating NPCs but not Müller glia because Rockout-treatment began after 28 h of light treatment. Our EdU/BrdU pulse-chase experiment revealed that fewer EdU-positive INL cells incorporated BrdU in Rock-inhibited retinas, indicating that decreased numbers of cells remained in the cell cycle. Because Rho/Rock signaling was shown previously to control the expression of cell-cycle-specific genes (Olson et al., 1995; Mammoto et al., 2004; Croft and Olson, 2006; Zhang et al., 2009), it might also regulate the decision to progress into another round of the cell cycle (Olson et al., 1995; Mammoto et al., 2004; Croft and Olson, 2006; Zhang et al., 2009). The recruitment of fewer Müller glia into the cell cycle in rock2 double morphants is consistent with Rock2a and Rock2b regulating Müller glia and possibly NPC proliferation.
Alternatively, the reduced number of proliferating cells could result from disrupted INM in the Rock-inhibited retina, rather than Rocks regulating the cell cycle directly. INM exposes nuclei to different spatially separated cues such as Notch-Delta signaling gradients that are implicated in regulating cell cycle exit in the developing retina (Murciano et al., 2002; Del Bene et al., 2008; Clark et al., 2012). However, inhibiting Notch signaling using the γ-secretase inhibitor R04929097, which is sufficient to recruit Müller glia into the cell cycle (Conner et al., 2014), did not maintain NPCs in the cell cycle in Rock-inhibited retinas, although more Müller glia were induced to proliferate. It is possible that Notch signaling plays different roles at distinct stages of retinal regeneration and that activation, rather than inhibition, of Notch signaling is required to maintain NPCs in the cell cycle.
The decreased numbers of rod and cone photoreceptors in Rock-inhibited retinas was likely due to a combination of factors: (1) reduced NPC proliferation, (2) increased cell death of ONL neurons at late stages of Rockout exposure, and (3) increased generation of ganglion/amacrine cells in the region between the rod photoreceptor nuclei and horizontal cell layer. The observed shift toward the production of ganglion/amacrine cells is similar to the phenotype observed in the INM-disrupted developing retina, where increased numbers of early born ganglion/amacrine cells were produced at the expense of later born neurons (Del Bene et al., 2008). However, in the developing retina, disrupting INM did not affect the position of these cells. In the regenerating Rockout-treated retina, the proliferating cells remaining in the ONL due to disrupted basal INM might be exposed to different concentrations of signaling factors that would typically prevent ganglion/amacrine cell formation in favor of rod and cone photoreceptors. GDF11, a member of the transforming growth factor-β superfamily, is one factor that is secreted to negatively regulate the generation of ganglion cells in the developing retina (Kim et al., 2005). Currently, it is unknown which signaling factors are expressed in the adult regenerating retina that shift the fate of NPCs toward those cells that were lost after light damage.
Mammals do not possess the endogenous capacity to regenerate lost retinal neurons. However, exogenous application of signaling molecules can induce limited Müller glia proliferation that produces low numbers of NPCs (Ooto et al., 2004; Osakada et al., 2007; Karl et al., 2008). The comparatively high endogenous regenerative capacity of adult zebrafish relative to the mammalian retina may correspond to the extent that Müller glia and NPCs undergo INM. Although mitotic cells were observed in the ONL of damaged mammalian retinas that were exposed to growth factors (Karl et al., 2008), it is unclear to what extent proliferating cells undergo INM and whether their early cell cycle exit/limited proliferation is due to ineffective INM.
Footnotes
This work was supported by the National Institutes of Health–National Eye Institute (Grant R01-EY018417 to D.R.H.) and the Center for Zebrafish Research, University of Notre Dame, Notre Dame, Indiana. We thank the Freimann Life Sciences technicians for care and husbandry of the zebrafish; members of the Hyde laboratory for thoughtful discussions; Ryne Gorsuch and Craig Nelson for input on retinal explant culture; Adam Henry and Adriaan de Feijter from Nikon Instruments for insight regarding multiphoton microscopy; William Archer and the Notre Dame Integrated Imaging Facility for support; and Betsy Garibaldi for help in sectioning retinal tissue.
The authors declare no competing financial interests.
References
- Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- Baye LM, Link BA. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J Neurosci. 2007;27:10143–10152. doi: 10.1523/JNEUROSCI.2754-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KA, Hart NH. Reorganization of filamentous actin and myosin-II in zebrafish eggs correlates temporally and spatially with cortical granule exocytosis. J Cell Sci. 1999;112:97–110. doi: 10.1242/jcs.112.1.97. [DOI] [PubMed] [Google Scholar]
- Bermel C, Tönges L, Planchamp V, Gillardon F, Weishaupt JH, Dietz GP, Bähr M, Lingor P. Combined inhibition of Cdk5 and ROCK additively increase cell survival, but not the regenerative response in regenerating retinal ganglion cells. Mol Cell Neurosci. 2009;42:427–437. doi: 10.1016/j.mcn.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Criquet P, Pantaloni D, Korn ED. Interaction of cytochalasin D with actin filaments in the presence of ADP and ATP. J Biol Chem. 1986;261:2041–2050. [PubMed] [Google Scholar]
- Cayouette M, Raff M. The orientation of cell division influences cell-fate choice in the developing mammalian retina. Development. 2003;130:2329–2339. doi: 10.1242/dev.00446. [DOI] [PubMed] [Google Scholar]
- Clark BS, Cui S, Miesfeld JB, Klezovitch O, Vasioukhin V, Link BA. Loss of Llgl1 in retinal neuroepithelia reveals links between apical domain size, Notch activity and neurogenesis. Development. 2012;139:1599–1610. doi: 10.1242/dev.078097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner C, Ackerman KM, Lahne M, Hobgood JS, Hyde DR. Repressing Notch signaling and expressing TNFalpha are sufficient to mimic retinal regeneration by inducing Müller glial proliferation to generate committed progenitor cells. J Neurosci. 2014;34:14403–14419. doi: 10.1523/JNEUROSCI.0498-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft DR, Olson MF. The Rho GTPase effector ROCK regulates cyclin A, cyclin D1, and p27Kip1 levels by distinct mechanisms. Mol Cell Biol. 2006;26:4612–4627. doi: 10.1128/MCB.02061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias TB, Yang YJ, Ogai K, Becker T, Becker CG. Notch signaling controls generation of motor neurons in the lesioned spinal cord of adult zebrafish. J Neurosci. 2012;32:3245–3252. doi: 10.1523/JNEUROSCI.6398-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsuch RA, Hyde DR. Regulation of Müller glial dependent neuronal regeneration in the damaged adult zebrafish retina. Exp Eye Res. 2014;123:131–140. doi: 10.1016/j.exer.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale IL, Fisher SK, Matsumoto B. The actin network in the ciliary stalk of photoreceptors functions in the generation of new outer segment discs. J Comp Neurol. 1996;376:128–142. doi: 10.1002/(SICI)1096-9861(19961202)376:1<128::AID-CNE8>3.0.CO;3B2-5. [DOI] [PubMed] [Google Scholar]
- Harding MJ, Nechiporuk AV. Fgfr-Ras-MAPK signaling is required for apical constriction via apical positioning of Rho-associated kinase during mechanosensory organ formation. Development. 2012;139:3130–3135. doi: 10.1242/dev.082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Haeseleer F, Hendrickson A. A comparison of immunocytochemical markers to identify bipolar cell types in human and monkey retina. Vis Neurosci. 2003;20:589–600. doi: 10.1017/S0952523803206015. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Herder C, Swiercz JM, Müller C, Peravali R, Quiring R, Offermanns S, Wittbrodt J, Loosli F. ArhGEF18 regulates RhoA-Rock2 signaling to maintain neuro-epithelial apico-basal polarity and proliferation. Development. 2013;140:2787–2797. doi: 10.1242/dev.096487. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, Pawson T, Jan Y, Stainier DY, Abdelilah-Seyfried S. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr Biol. 2001;11:1492–1502. doi: 10.1016/S0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- Hutchison C, Kill I. Changes in the nuclear distribution of DNA polymerase alpha and PCNA/cyclin during the progress of the cell cycle, in a cell-free extract of Xenopus eggs. J Cell Sci. 1989;93:605–613. doi: 10.1242/jcs.93.4.605. [DOI] [PubMed] [Google Scholar]
- Ikebe M, Koretz J, Hartshorne DJ. Effects of phosphorylation of light chain residues threonine 18 and serine 19 on the properties and conformation of smooth muscle myosin. J Biol Chem. 1988;263:6432–6437. [PubMed] [Google Scholar]
- Karl MO, Hayes S, Nelson BR, Tan K, Buckingham B, Reh TA. Stimulation of neural regeneration in the mouse retina. Proc Natl Acad Sci U S A. 2008;105:19508–19513. doi: 10.1073/pnas.0807453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen SC, Ramanan V, Montgomery JE, T Burket C, Liu CG, Vihtelic TS, Hyde DR. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67:1009–1031. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308:1927–1930. doi: 10.1126/science.1110175. [DOI] [PubMed] [Google Scholar]
- Koch JC, Tönges L, Barski E, Michel U, Bähr M, Lingor P. ROCK2 is a major regulator of axonal degeneration, neuronal death and axonal regeneration in the CNS. Cell Death Dis. 2014;5:e1225. doi: 10.1038/cddis.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y, Suetsugu T, Suda M, Mimori-Kiyosue Y, Toida K, Baba SA, Kimura A, Matsuzaki F. Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. EMBO J. 2011;30:1690–1704. doi: 10.1038/emboj.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroehne V, Freudenreich D, Hans S, Kaslin J, Brand M. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development. 2011;138:4831–4841. doi: 10.1242/dev.072587. [DOI] [PubMed] [Google Scholar]
- Lee HO, Norden C. Mechanisms controlling arrangements and movements of nuclei in pseudostratified epithelia. Trends Cell Biol. 2013;23:141–150. doi: 10.1016/j.tcb.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Leung L, Klopper AV, Grill SW, Harris WA, Norden C. Apical migration of nuclei during G2 is a prerequisite for all nuclear motion in zebrafish neuroepithelia. Development. 2011;138:5003–5013. doi: 10.1242/dev.071522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GP, Matsumoto B, Fisher SK. Changes in the organization and expression of cytoskeletal proteins during retinal degeneration induced by retinal detachment. Invest Ophthalmol Vis Sci. 1995;36:2404–2416. [PubMed] [Google Scholar]
- Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- Mammoto A, Huang S, Moore K, Oh P, Ingber DE. Role of RhoA, mDia, and ROCK in cell shape-dependent control of the Skp2–p27kip1 pathway and the G1/S transition. J Biol Chem. 2004;279:26323–26330. doi: 10.1074/jbc.M402725200. [DOI] [PubMed] [Google Scholar]
- Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S, Tsukita S. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J Cell Biol. 1998;140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EJ, Ikmi A, Gibson MC. Interkinetic nuclear migration is a broadly conserved feature of cell division in pseudostratified epithelia. Curr Biol. 2011;21:485–491. doi: 10.1016/j.cub.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Montgomery JE, Parsons MJ, Hyde DR. A novel model of retinal ablation demonstrates that the extent of rod cell death regulates the origin of the regenerated zebrafish rod photoreceptors. J Comp Neurol. 2010;518:800–814. doi: 10.1002/cne.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murciano A, Zamora J, López-Sánchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- Nagashima M, Barthel LK, Raymond PA. A self-renewing division of zebrafish Müller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013;140:4510–4521. doi: 10.1242/dev.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Suzuki Y, Kihira H, Wada H, Fujioka M, Ito M, Nakano T, Kaibuchi K, Shiku H, Nishikawa M. Regulation of myosin phosphatase through phosphorylation of the myosin-binding subunit in platelet activation. Blood. 1997;90:3936–3942. [PubMed] [Google Scholar]
- Nelson CM, Gorsuch RA, Bailey TJ, Ackerman KM, Kassen SC, Hyde DR. Stat3 defines three populations of Müller glia and is required for initiating maximal Müller glia proliferation in the regenerating zebrafish retina. J Comp Neurol. 2012;520:4294–4311. doi: 10.1002/cne.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Ackerman KM, O'Hayer P, Bailey TJ, Gorsuch RA, Hyde DR. Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Müller glia proliferation during zebrafish retinal regeneration. J Neurosci. 2013;33:6524–6539. doi: 10.1523/JNEUROSCI.3838-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Namba T, Shinoda T, Kondo T, Watanabe T, Inoue Y, Takeuchi K, Enomoto Y, Ota K, Oda K, Wada Y, Sagou K, Saito K, Sakakibara A, Kawaguchi A, Nakajima K, Adachi T, Fujimori T, Ueda M, Hayashi S, et al. TAG-1-assisted progenitor elongation streamlines nuclear migration to optimize subapical crowding. Nat Neurosci. 2013;16:1556–1566. doi: 10.1038/nn.3525. [DOI] [PubMed] [Google Scholar]
- Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- Ooto S, Akagi T, Kageyama R, Akita J, Mandai M, Honda Y, Takahashi M. Potential for neural regeneration after neurotoxic injury in the adult mammalian retina. Proc Natl Acad Sci U S A. 2004;101:13654–13659. doi: 10.1073/pnas.0402129101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F, Ooto S, Akagi T, Mandai M, Akaike A, Takahashi M. Wnt signaling promotes regeneration in the retina of adult mammals. J Neurosci. 2007;27:4210–4219. doi: 10.1523/JNEUROSCI.4193-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson RA, Lüneborg NL, Becker DL, Mobbs P. Gap junctions modulate interkinetic nuclear movement in retinal progenitor cells. J Neurosci. 2005;25:10803–10814. doi: 10.1523/JNEUROSCI.2312-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggi L, Vitorino M, Masai I, Harris WA. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J Cell Biol. 2005;171:991–999. doi: 10.1083/jcb.200509098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk J, Wilsch-Bräuninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci U S A. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear PC, Erickson CA. Apical movement during interkinetic nuclear migration is a two-step process. Dev Biol. 2012;370:33–41. doi: 10.1016/j.ydbio.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surzenko N, Crowl T, Bachleda A, Langer L, Pevny L. SOX2 maintains the quiescent progenitor cell state of postnatal retinal Müller glia. Development. 2013;140:1445–1456. doi: 10.1242/dev.071878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JL, Ochocinska MJ, Hitchcock PF, Thummel R. Using the Tg(nrd:egfp)/albino zebrafish line to characterize in vivo expression of neurod. PLoS One. 2012;7:e29128. doi: 10.1371/journal.pone.0029128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Kassen SC, Montgomery JE, Enright JM, Hyde DR. Inhibition of Müller glial cell division blocks regeneration of the light-damaged zebrafish retina. Dev Neurobiol. 2008a;68:392–408. doi: 10.1002/dneu.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Kassen SC, Enright JM, Nelson CM, Montgomery JE, Hyde DR. Characterization of Müller glia and neuronal progenitors during adult zebrafish retinal regeneration. Exp Eye Res. 2008b;87:433–444. doi: 10.1016/j.exer.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Enright JM, Kassen SC, Montgomery JE, Bailey TJ, Hyde DR. Pax6a and Pax6b are required at different points in neuronal progenitor cell proliferation during zebrafish photoreceptor regeneration. Exp Eye Res. 2010;90:572–582. doi: 10.1016/j.exer.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda S, Kitagawa T, Takashima S, Asakawa S, Shimizu N, Mitani H, Shima A, Tsutsumi M, Hori H, Naruse K, Ishikawa Y, Takeda H. FAK-mediated extracellular signals are essential for interkinetic nuclear migration and planar divisions in the neuroepithelium. J Cell Sci. 2010;123:484–496. doi: 10.1242/jcs.057851. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::AID-NEU1>3.0.CO;3B2-H. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Doro CJ, Hyde DR. Cloning and characterization of six zebrafish photoreceptor opsin cDNAs and immunolocalization of their corresponding proteins. Vis Neurosci. 1999;16:571–585. doi: 10.1017/s0952523899163168. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Soverly JE, Kassen SC, Hyde DR. Retinal regional differences in photoreceptor cell death and regeneration in light-lesioned albino zebrafish. Exp Eye Res. 2006;82:558–575. doi: 10.1016/j.exer.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Wan J, Ramachandran R, Goldman D. HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev Cell. 2012;22:334–347. doi: 10.1016/j.devcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Cadwallader AB, Jang DS, Tsang M, Yost HJ, Amack JD. The Rho kinase Rock2b establishes anteroposterior asymmetry of the ciliated Kupffer's vesicle in zebrafish. Development. 2011;138:45–54. doi: 10.1242/dev.052985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber IP, Ramos AP, Strzyz PJ, Leung LC, Young S, Norden C. Mitotic position and morphology of committed precursor cells in the zebrafish retina adapt to architectural changes upon tissue maturation. Cell Rep. 2014;7:386–397. doi: 10.1016/j.celrep.2014.03.014. [DOI] [PubMed] [Google Scholar]
- Wei X, Luo Y, Hyde DR. Molecular cloning of three zebrafish lin7 genes and their expression patterns in the retina. Exp Eye Res. 2006;82:122–131. doi: 10.1016/j.exer.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Xie Z, Moy LY, Sanada K, Zhou Y, Buchman JJ, Tsai LH. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Maruyama K, Himori N, Omodaka K, Yokoyama Y, Shiga Y, Morin R, Nakazawa T. The novel Rho kinase (ROCK) inhibitor K-115: a new candidate drug for neuroprotective treatment in glaucoma. Invest Ophthalmol Vis Sci. 2014;55:7126–7136. doi: 10.1167/iovs.13-13842. [DOI] [PubMed] [Google Scholar]
- Yarrow JC, Totsukawa G, Charras GT, Mitchison TJ. Screening for cell migration inhibitors via automated microscopy reveals a Rho-kinase inhibitor. Chem Biol. 2005;12:385–395. doi: 10.1016/j.chembiol.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Yu J, Lei K, Zhou M, Craft CM, Xu G, Xu T, Zhuang Y, Xu R, Han M. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Tang Q, Xu F, Xue Y, Zhen Z, Deng Y, Liu M, Chen J, Liu S, Qiu M, Liao Z, Li Z, Luo D, Shi F, Zheng Y, Bi F. RhoA regulates G1-S progression of gastric cancer cells by modulation of multiple INK4 family tumor suppressors. Mol Cancer Res. 2009;7:570–580. doi: 10.1158/1541-7786.MCR-08-0248. [DOI] [PubMed] [Google Scholar]