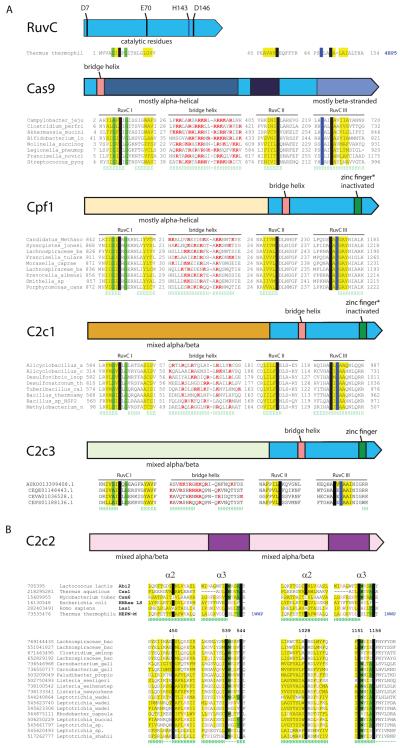
Figure 2. Domain architectures and conserved motifs of the Class 2 effector proteins.
(A) Types II and V: TnpB-derived nucleases. The top panel shows the RuvC nuclease from Thermus thermophilus (PDB ID: 4EP5) with the catalytic amino acid residues denoted. Underneath each domain architecture, an alignment of the conserved motifs in selected representatives of the respective protein family (a single sequence for RuvC) is shown. The catalytic residues are shown by white letters on a black background; conserved hydrophobic residues are highlighted in yellow; conserved small residues are highlighted in green; in the bridge helix alignment, positively charged residues are in red. Secondary structure prediction is shown underneath the aligned sequences: H denotes α-helix and E denotes extended conformation (β-strand). The poorly conserved spacers between the alignment blocks are shown by numbers. See also Figures S3, S4 and S5.
(B) Type VI: predicted RNases containing two HEPN domains. The top alignment blocks include selected HEPN domains described previously and the bottom blocks include the catalytic motifs from the putative type VI effector proteins. The designations are as in (A). See also Figures S3 and S6.