Analysis of a new structure of an Epsilon class glutathione transferase from Drosophila melanogaster reveals a highly conserved motif that spans the dimeric subunit interface and connects the two active sites.
Keywords: dimer interface, epsilon crystal structure, GSTE6, insect glutathione transferase, protein structure
Abstract
Epsilon class glutathione transferases (GSTs) have been shown to contribute significantly to insecticide resistance. We report a new Epsilon class protein crystal structure from Drosophila melanogaster for the glutathione transferase DmGSTE6. The structure reveals a novel Epsilon clasp motif that is conserved across hundreds of millions of years of evolution of the insect Diptera order. This histidine-serine motif lies in the subunit interface and appears to contribute to quaternary stability as well as directly connecting the two glutathiones in the active sites of this dimeric enzyme.
INTRODUCTION
Glutathione transferases (GSTs) are a superfamily of proteins found in all cellular organisms, which usually exist as multiple isoforms [1,2]. This extensive family has been categorized into at least 13 classes. Four classes, Omega, Sigma, Theta and Zeta, are found in all or most metazoans. There are also classes that appear to be found only in specific organisms such as in plants, Phi and Tau [3], or in arthropods, Delta and Epsilon [4]. One general function of this superfamily is the involvement in detoxication reactions for both endogenous and xenobiotic compounds. Other specific physiological roles have been identified for individual enzymes; in mammals the Sigma GST has been shown to be prostaglandin D2 synthase [5]; and in Drosophila an Omega GST was found to be a key synthase enzyme in pteridine biosynthesis of red eye pigments [6].
In insects, the Delta and Epsilon class GSTs make major contributions to detoxication of insecticides, contributing to insecticide resistance [7,8]. Currently, there are seven Epsilon structures available; allelic proteins with different ligands from the mosquito malaria vector Anopheles gambiae [9,10], two allelic proteins from a second major mosquito malaria vector Anopheles funestus [11] and a structure from the housefly Musca domestica [12]. Therefore, additional structural data would increase our understanding of this enzyme class.
In the present study, we present a novel crystal structure of an Epsilon GST (DmGSTE6) from Drosophila melanogaster. DmGSTE6 has significantly high activity for 4-hydroxynonenal (HNE) [13]. HNE was first hypothesized to be a secondary cytotoxic end product of lipid peroxidation that contributes to the aetiology of degenerative disorders like Alzheimer's [14] and Parkinson's disease [15]. HNE is now known to also be a concentration dependent signalling molecule that modulates several pathways [16]. GSTs may therefore regulate the HNE modulation of these signalling pathways. The gene encoding DmGSTE6 is of interest as it originates from a template gene, which during the course of Drosophila evolution has generated several Epsilon genes in the Epsilon gene cluster [17]. Within the DmGSTE6 structure we have identified a motif that has been conserved across millions of years of evolution. This motif lies at the interface extending into each subunit and is similar to, but more complex than, the interface motif called the ‘clasp’ motif previously reported for Delta GSTs [18]. Residues from the Epsilon motif also reside in the active site and are likely to contribute to an electron-sharing network [19].
MATERIALS AND METHODS
Production and purification of DmGSTE6
The DNA encoding DmGSTE6 was engineered into a pET-21d(+) (Novagen) plasmid and transformed into Escherichia coli BL21 (DE3) cells. Overnight stationary-phase cultures in Luria–Bertani (LB) broth (8 ml) were used to inoculate 800 ml LB broth, both supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. The cells were allowed to grow for 3 h at 310 K and then were cooled to 291 K and induced with 0.1 mM β-D-1-thiogalactopyranoside (IPTG) and incubated overnight for 16 h. The cultures were harvested at 7000 × g for 10 min and the pellets were kept at 253 K until used. The induced culture pellet was suspended with 20 ml of PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3) containing 400 μl of 100 mg/ml lysozyme, 10 mM DTT and 10 μl of 1.4 M β-mercaptoethanol by gentle vortex-mixing. The cell suspension was incubated on ice for 20 min and the crude cell lysate was obtained by sonication. The lysed cells were centrifuged at 10000 × g and 277 K for 30 min. The soluble DmGSTE6 in the supernatant fraction was purified using a GSTrap FF column. The 5 ml column was equilibrated with five column volumes (CV) of PBS buffer. The supernatant was applied to the column with a flow rate of 5 ml/min. The non-specific binding proteins were eluted with 10 CV of PBS buffer. The bound GST was eluted with five CV of elution buffer (10 mM GSH, 50 mM Tris–HCl, pH 8.0, 10 mM DTT).
Crystallization
DmGSTE6 at 15–20 mg/ml was crystallized in the presence of 10 mM GSH under a condition containing 20% PEG3350 and 0.2 M sodium thiocyanate in a 1:1 ratio by sitting drop vapour diffusion at 288 K. Protein crystals appeared within 3 days.
Data collection and processing
X-ray diffraction data were collected on beamline I03 at Diamond Light Source (DLS) with an X-ray wavelength of 0.976 Å (1 Å=0.1 nm), to a resolution of 1.72 Å. The crystal was oscillated in the beam by 0.1° per frame over a range of 360°. Data were processed and scaled using xia2 [20] and AIMLESS from the CCP4 suite [21]. Initial phases were obtained by molecular replacement with PHASER [22] using a GST Epsilon class from A. gambiae (PDB: 2IMI) as the search model. Model building was performed with COOT [23] and restrained refinement with REFMAC in CCP4 [21] and PHENIX [24]. The data collection and refinement statistics are shown in Table 1. Molecular graphics and analyses were performed with the UCSF Chimera package [25] and PyMol (The PyMol Molecular Graphic System, version 1.5.0.3 Schrödinger, LLC). Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). The atomic coordinates of the crystal structure have been deposited into the Protein Data Bank (PDB access code 4YH2).
Table 1. Data collection and refinement statistics for GSTE6.
*Highest resolution shell is shown in parenthesis.
| Data collection | |
|---|---|
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 176.4, 58.9, 122.8 |
| α, β, γ (°) | 90.0, 128.2, 90.0 |
| Resolution (Å) | 96.4–1.72 (1.76–1.72) |
| Rmerge | 0.09 (0.69) |
| I/σI | 12.3 (2.4) |
| Completeness (%) | 99.8 (99.9) |
| Redundancy | 6.6 (6.9) |
| Refinement | |
| Resolution (Å) | 96.4–1.72 (1.74–1.72) |
| No. reflections | 104998 (3266) |
| Rwork/Rfree | 0.157 (0.272)/0.188 (0.357) |
| No. atoms | 8099 |
| Protein | 7120 |
| Water | 979 |
| B-factors (Å2) | |
| Protein | 24.3 |
| Water | 34.6 |
| RMSDs | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.057 |
| Ramachandran | |
| Favoured (%) | 98.1 |
| Allowed (%) | 1.7 |
| Outliers (%) | 0.2 |
Sequence alignment
Sequence alignment is done by Clustal X, version 2.0.10 [26] and displayed with GeneDoc (http://www.psc.edu/biomed/genedoc).
RESULTS AND DISCUSSION
In the present study, we have determined the structure of DmGSTE6 to supplement the currently known Epsilon structures (Figure 1, Table 1). This structure adopts the typically highly conserved GST fold with two domains comprising a βαβαββα motif in the N-terminus and an all α-helical domain in the C-terminus.
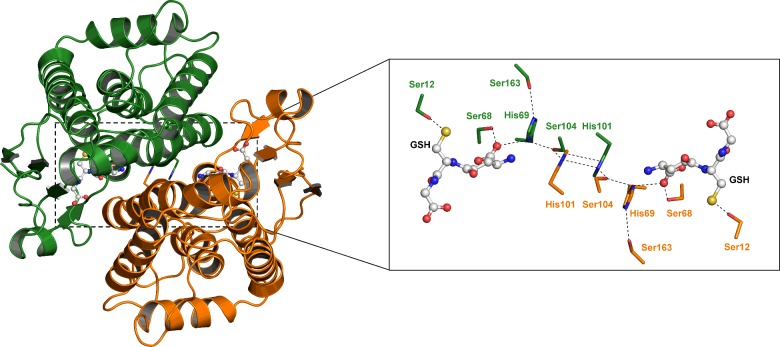
Figure 1. Ribbon representation of the D. melanogaster GSTE6 dimer.
Subunits (A) and (B) are distinguished by different colours, green and orange. The inset shows the ‘wafer’ histidine interface motif accompanied by serine residues connecting the two subunits from the active site of one subunit to the other. Ser12 is the catalytic residue.
Previously, we have carried out structure–function studies on Delta class GSTs, which identified an interface lock-and-key ‘clasp’ motif that is structurally conserved within the Delta class enzymes [18]. Amino acid sequence alignments predict that this motif may also be conserved in the Epsilon class GSTs. The structural motif appears to affect protein dynamics and therefore influences substrate specificity, enzyme activity and protein stability. Analysis of the clasp formation in this Epsilon DmGSTE6 reveals that the interface motif is more complex than observed for the Delta class. The Epsilon clasp motif comprises an extended ‘wafer’ arrangement of four histidines (two contributed from each subunit), which is supported by interactions with several conserved serines from helices 3, 4 and 6 (Figure 1). This elongated motif stretches across the interface deep into both subunits of the homodimer. His101 from one subunit wraps around the His101 from the other subunit to generate the clasp motif also seen in the Delta GST class [18]. This arrangement of the histidines involves aromatic ring stacking and pi–pi interaction of the two residues. In Delta class GSTs, the clasp motif has been shown to stabilize the quaternary structure as well as have a role in subunit communication between active sites [18]. The structural contributions of the clasp motif in the Delta GST also have an impact on catalytic specificity and the efficiency of the enzyme. In the Epsilon DmGSTE6, a second histidine in each subunit, His69, interacts with clasp His101 from the other subunit (Figure 1). These His–His interactions are also supported by three serines, Ser68, Ser104, Ser163, in each subunit (Figure 1). This extended motif would appear to play a structural role, as it is internal and spans the subunit interface. Furthermore, Ser68 directly interacts with GSH in the active site as the Ser68 OG and N atoms are within 2.5–3.3 Å of the GSH O11 and O12 atoms (Figure 1). The His69 ND1 atom is also within the interaction distance of 3.5 Å of the GSH O12 atom. Both Ser68 and His69 are located at the N-terminus of alpha helix 3 and possibly exploit the helix dipole to strengthen these interactions. Residues Ser68 and His69 in DmGSTE6 are the equivalent residues to Ser65 and Arg66, in the active site of the Delta class AdGSTD3, which are reported to be components of an electron-sharing network utilized for catalysis [19,27,28]. An amino acid sequence alignment of the six Epsilon GSTs, for which structures are available, is shown in Figure 2. The His wafer residues are conserved in these Epsilon class proteins and superposition of their structures shows that this sequence conservation translates into side chain position conservation in their 3D structures (Figure 3).
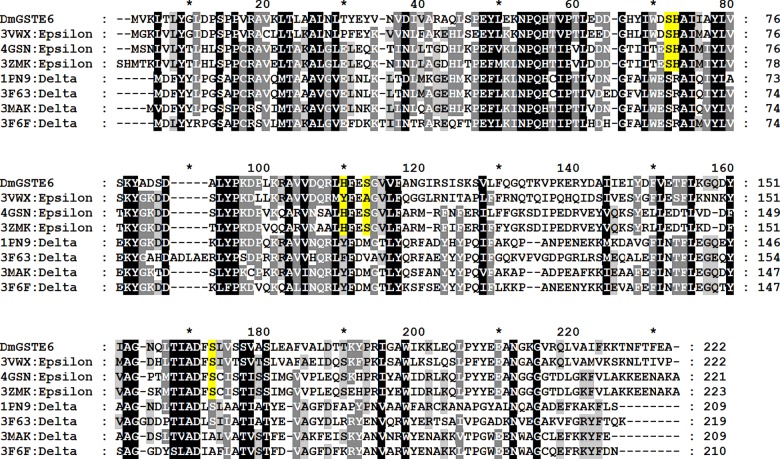
Figure 2. Amino acid alignment of the Epsilon GSTs compared with Delta GSTs with available crystal structures.
Highlighted in yellow is the conserved histidine wafer interface motif consisting of two histidines and three supporting serines from each subunit. DmGSTE6 in the present study refers to D. melanogaster GST Epsilon 6. The PDB ID codes are: 3VWX M. domestica Epsilon; 4GSN A. gambiae Epsilon 2; 3ZMK A. funestus Epsilon 2; 1PN9 A. gambiae Delta 1; 3F63 Anopheles dirus Delta 4; 3MAK D. melanogaster Delta 1; 3F6F D. melanogaster Delta 10.
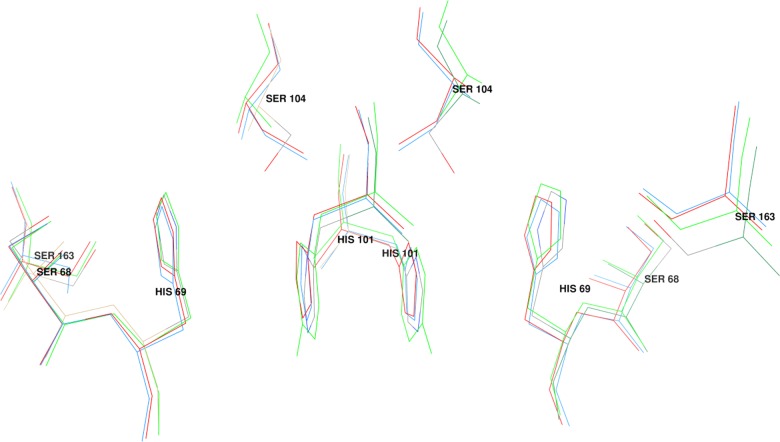
Figure 3. Superposition of the histidine interface motif of Epsilon GSTs from four species.
The residue numbering refers to the D. melanogaster GST Epsilon 6. The superimposed structures are 4YH2 (the present study, D. melanogaster), 2IMI (A. gambiae), 3ZML (A. funestus) and 3VWX (M. domestica).
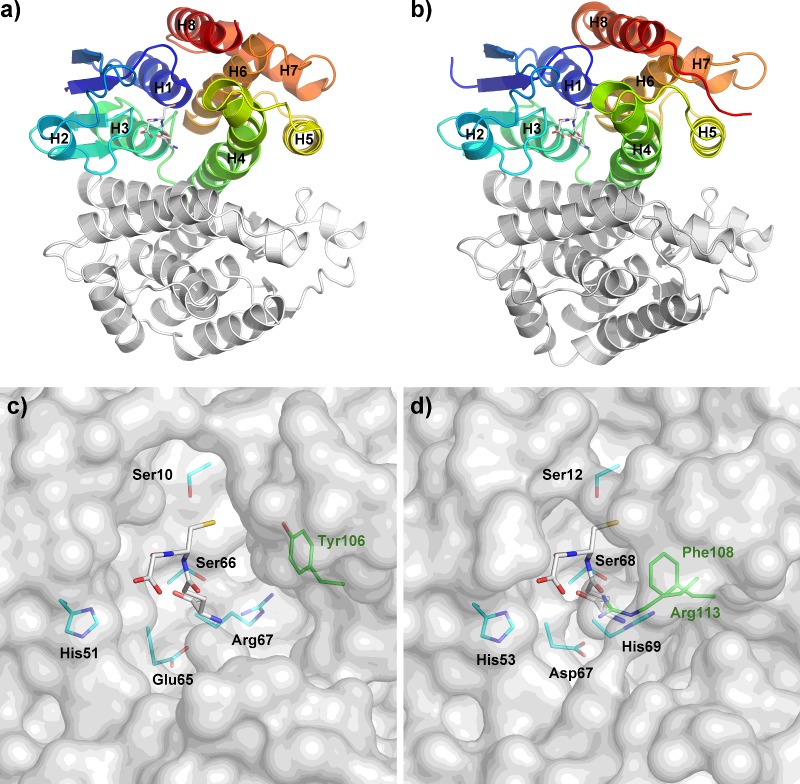
The current structure also shows that the active site pocket for Epsilon class GSTs is more in a ‘closed’ configuration than that of Delta class. Major contributions to this difference arise from the arrangement of helices, especially the positioning of alpha helix 4 (Figures 4a and 4b). This offers an extra GSH interaction with Arg113 from the C-terminal domain, which is a novel feature and perhaps is unique to Epsilon class GSTs (Figures 4c and 4d). To validate the conservation of this arginine and other GSH-binding residues, all Epsilon and Delta class GSTs were retrieved from complete genomes of D. melanogaster and A. gambiae for protein sequence analysis. Based on previous annotation, this includes 14 Epsilon/12 Delta class GSTs from D. melanogaster [13] and 8 Epsilon/15 Delta class GSTs from A. gambiae [29]. Besides the highly conserved catalytic serine (Ser12), we categorized GSH-binding residues into three different groups namely the GSH glycyl moiety, the GSH glutamyl moiety and the C-terminal domain interactor (Figure 5). The impact of protein–ligand interactions toward the glycyl and glutamyl regions of GSH have been extensively studied for the Delta class GSTs. Amino acid conservation in these positions has been reported to play a significant role in enzyme catalysis, substrate specificity and structural integrity of the enzyme [19,27,28,30]. The histidine that interacts with the glycyl moiety of GSH (His53 in DmGSTE6) is highly conserved in both Delta and Epsilon classes of GSTs (Figure 5a). Amino acid replacement of the equivalent residue to His53 in a Delta class GST exhibits a catalytic efficiency approximately 5200-fold lower than observed for the wild-type [28]. For the GSH glutamyl moiety (Figure 5b), this region of interactions also represents functionally conserved residues in both GST classes, and influences protein folding, enzyme kinetics, the rate-limiting step, GSH ionization and the electron-sharing network in catalysis [19,27,30].). It is of interest that the strictly conserved His69 in Epsilon class, which is involved in GSH glutamyl binding, is also one of the ‘wafer’ histidines in the interface motif. This position has a significant role in GSH-binding affinity, substrate specificity and protein folding. As a result, His69, which links the subunits across the dimeric interface and communicates from one active site to the other, may be an important regulating residue for the Epsilon class of GSTs. The final pair of GSH-binding residues in Epsilon class GSTs lies in the C-terminal domain, Phe106 and Arg113. This region is of particular interest since it lies on alpha helix 4, which is positioned relatively closer to the N-terminal domain compared with the Delta class GSTs. Arg113 is conserved only in the Epsilon class (Figure 5c) and it interacts with the glutamyl part of GSH, suggesting that this residue may play a role in modulating GSH/hydrophobic substrate conjugation. Additionally, structural comparison focusing on alpha helix 4 also shows that Phe108 in GSTE6, though conserved in both insect-specific classes of GST (Figure 2), is close enough for hydrophobic interaction with GSH only in the Epsilon class. Distance from Phe108 CE1 atom of DmGSTE6 to GSH CB1 is 3.5 Å whereas the closest distance between dmGSTD1 and GSH is 5.6 Å from Tyr106 CE2 atom to GSH OE1 atom.
Figure 4. Active site pockets of DmGSTE6 in comparison with dmGSTD1.
Arrangement of helices in dmGSTD1 (a and c) and DmGSTE6 (b and d) contributes to the shape of active site pockets. Stick representation of glutathione is in white and its interacting residues from the N-terminal and the C-terminal domain are in cyan and green, respectively. Tyr106 in panel c is for comparison only.
Figure 5. Graphical representation of amino acid multiple sequence alignment for glutathione binding residues of Epsilon and Delta class GSTs from D. melanogaster and A. gambiae genomes.
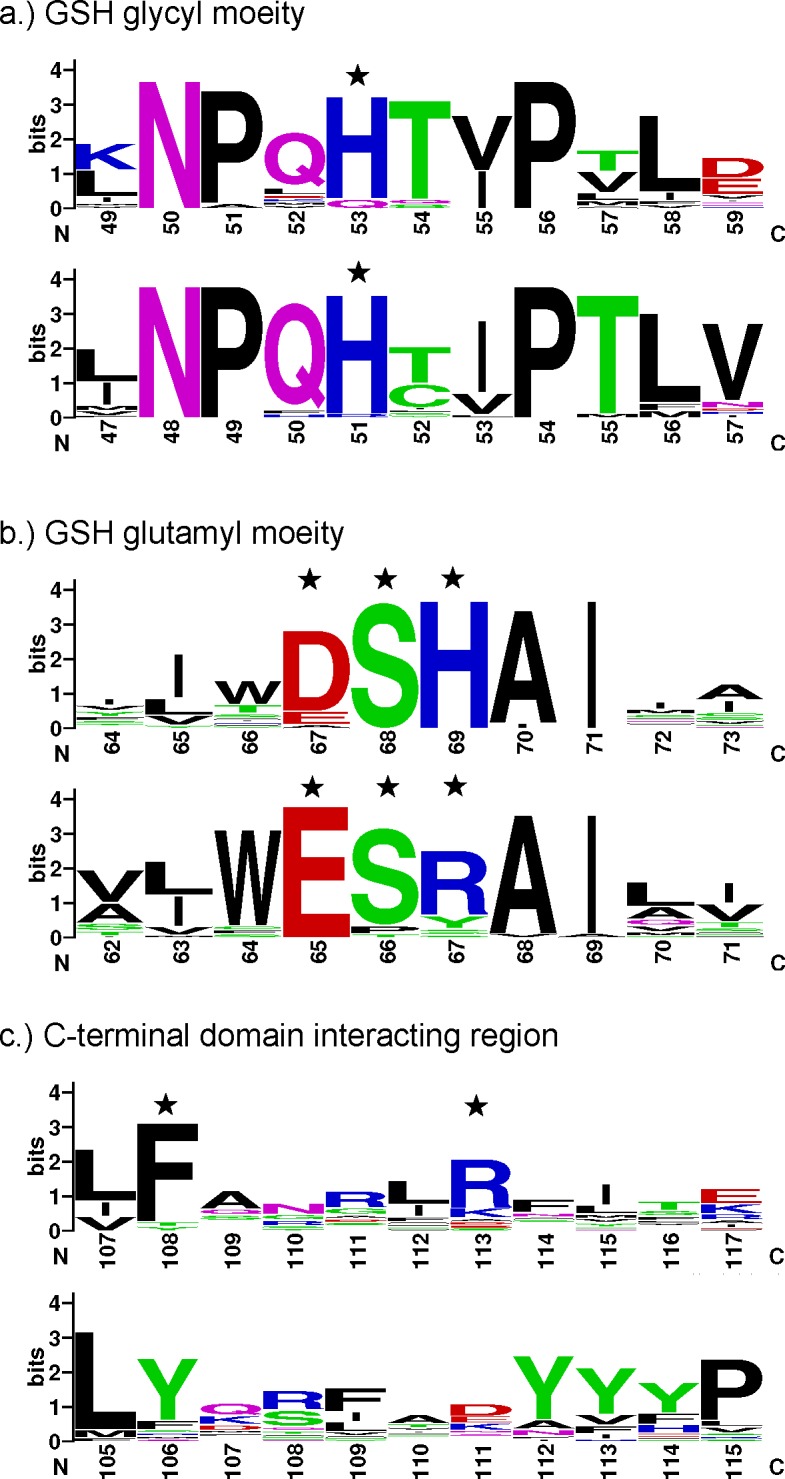
The glutathione interacting residues are indicated with stars. The top panel in each category represents Epsilon class GSTs and the bottom panel represents Delta class GSTs with respective residue numbers on an X-axis. The image was generated by WebLogo [44,45]. A logo represents each column of the alignment by a stack of letters, with the height of each letter proportional to the observed frequency of the corresponding amino acid, and the overall height of each stack proportional to the sequence conservation at that position [45]. The letters of each stack are ordered from most to least frequent, so that one may read the consensus sequence from the tops of the stacks [45]. Single letter amino acid symbols are coloured according to their chemical properties; polar amino acids (G, S, T, Y, C, Q, N) are green, basic (K, R, H) blue, acidic (D and E) red, and hydrophobic (A, V, L, I, P, W, F, M) amino acids are black [45].
As stated above, in D. melanogaster DmGSTE6 has significantly high activity for HNE although not as great as reported for human GSTA4 [31], it is greater than what has been observed for two other human Alpha GSTs, GSTA1 and GSTA2 [32]. Therefore superposition of the structure of human Alpha class GSTA4 with GSH–HNE conjugate analogue in the active site (PDB ID: 3IK7; [33]) and the DmGSTE6 allowed us to identify possible residues involved in HNE binding. In GSTA4 these residues have been identified as Tyr9 (catalytic residue); Arg15 (N-terminus of helix 1); Ile107, Met108, Phe111 (helix 4); Tyr212, Val216, Tyr217, Phe220 (helix 8) [33]. From superposition the residues for DmGSTE6 that would generate a hydrophobic binding cavity for HNE appear to be Ser12 (catalytic residue); Pro11, Pro13, Pro14 (N-terminus helix 1); Ile36 (coil between beta strand 2 and helix 2); Phe108, Ile112, Arg113, Leu120 (helix 4); Phe174 (C-terminus helix 7); Leu209, Phe213 (helix 9). Although appearing to generate a similar hydrophobic cavity the residues of DmGSTE6 originate from different regions of the protein. This suggests convergent evolutionary pressure occurred in the Epsilon and Alpha GST classes to independently obtain the HNE activity.
Both Delta and Epsilon class GSTs have been shown to be important for contributing to insecticide resistance in mosquito malaria and dengue vectors [9–11,34]. At present, both GST classes, Delta and Epsilon, appear to be found only in arthropods. Delta GSTs have been reported in the arthropod Sarcoptes scabiei, a parasitic arachnid [35], a copepod crustacean Tigriopus japonicus [36] and the Chinese mitten crab Eriocheir sinensis [37]. Genomes from the four major insect orders, Coleoptera (beetle), Diptera (true flies), Hymenoptera (wasps, ants and bees) and Lepidoptera (moths and butterflies), have been reported to possess Delta GSTs [38]. In contrast, the current fully sequenced Hymenoptera genomes (Apis mellifera and Nasonia vitripennis) lack the Epsilon class, which suggests that Epsilon class GSTs appeared after the Hymenoptera divergence, approximately 238.5–307.2 million years ago [39,40]. The currently available Epsilon structures are from species of the Diptera order which is thought to have rapidly radiated during the Triassic period, 201–252 million years ago [41–43]. As a frame of reference, it is estimated that humans and mice diverged 62–100 million years ago [39]. Thus the Epsilon GST class has been evolving for a significant period of time, while maintaining this distinct Epsilon clasp structural motif in species across the Diptera order. This suggests that the Epsilon clasp motif serves a critical function, possibly in structure maintenance and in contributing to catalysis through the electron-sharing network.
Postscript
During the writing of this manuscript, two newly deposited structures of Drosophila GSTE6 (4PNF) and GSTE7 (4PNG) appeared in the PDB. This DmGSTE6 has structural similarity to the one in the present study (4YH2) with RMSD of 0.24 Å including 2840 atoms.
Acknowledgments
We thank the Diamond Light Source (proposal MX8423) for synchrotron beamtime on beamline I03, and Jonathan Grimes for help with data collection.
Abbreviations
- CV
column volumes
- DmGSTE6
Drosophila melanogaster glutathione transferase epsilon 6
- GST
glutathione transferase
- HNE
4-hydroxynonenal
- LB
Luria–Bertani
AUTHOR CONTRIBUTION
Jantana Wongsantichon performed the experiments. Jantana Wongsantichon and Albert J. Ketterman analysed the data. Jantana Wongsantichon, Robert C. Robinson and Albert J. Ketterman contributed to the manuscript preparation.
FUNDING
This work was supported by the Office of Higher Education Commission, Mahidol University under the National Research Universities Initiative and Mahidol University and the Agency for Science, Technology and Research (A*STAR), Singapore (to J.W. and R.C.R.).
References
- 1.Ketterer B. A bird's eye view of the glutathione transferase field. Chem. Biol. Interact. 2001;138:27–42. doi: 10.1016/S0009-2797(01)00277-0. [DOI] [PubMed] [Google Scholar]
- 2.Hayes J.D., Flanagan J.U., Jowsey I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 3.Edwards R., Dixon D.P. Plant glutathione transferases. Methods Enzymol. 2005;401:169–186. doi: 10.1016/S0076-6879(05)01011-6. [DOI] [PubMed] [Google Scholar]
- 4.Ketterman A.J., Saisawang C., Wongsantichon J. Insect glutathione transferases. Drug Metab. Rev. 2011;43:253–265. doi: 10.3109/03602532.2011.552911. [DOI] [PubMed] [Google Scholar]
- 5.Flanagan J.U., Smythe M.L. Sigma-class glutathione transferases. Drug Metab. Rev. 2011;43:194–214. doi: 10.3109/03602532.2011.560157. [DOI] [PubMed] [Google Scholar]
- 6.Kim J., Suh H., Kim S., Kim K., Ahn C., Yim J. Identification and characteristics of the structural gene for the Drosophila eye color mutant sepia, encoding PDA synthase, a member of the Omega class glutathione S-transferases. Biochem. J. 2006;398:451–460. doi: 10.1042/BJ20060424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranson H., Rossiter L., Ortelli F., Jensen B., Wang X., Roth C.W., Collins F.H., Hemingway J. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochem. J. 2001;359:295–304. doi: 10.1042/bj3590295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortelli F., Rossiter L.C., Vontas J., Ranson H., Hemingway J. Heterologous expression of four glutathione transferase genes genetically linked to a major insecticide-resistance locus from the malaria vector Anopheles gambiae. Biochem. J. 2003;373:957–963. doi: 10.1042/bj20030169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Qiu L., Ranson H., Lumjuan N., Hemingway J., Setzer W.N., Meehan E.J., Chen L. Structure of an insect epsilon class glutathione S-transferase from the malaria vector Anopheles gambiae provides an explanation for the high DDT-detoxifying activity. J. Struct. Biol. 2008;164:228–235. doi: 10.1016/j.jsb.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell S.N., Rigden D.J., Dowd A.J., Lu F., Wilding C.S., Weetman D., Dadzie S., Jenkins A.M., Regna K., Boko P., et al. Metabolic and target-site mechanisms combine to confer strong DDT resistance in Anopheles gambiae. PLoS One. 2014;9:e92662. doi: 10.1371/journal.pone.0092662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riveron J.M., Yunta C., Ibrahim S.S., Djouaka R., Irving H., Menze B.D., Ismail H.M., Hemingway J., Ranson H., Albert A., Wondji C.S. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 2014;15:R27. doi: 10.1186/gb-2014-15-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura C., Yajima S., Miyamoto T., Sue M. Structural analysis of an epsilon-class glutathione transferase from housefly, Musca domestica. Biochem. Biophys. Res. Commun. 2013;430:1206–1211. doi: 10.1016/j.bbrc.2012.12.077. [DOI] [PubMed] [Google Scholar]
- 13.Saisawang C., Wongsantichon J., Ketterman A.J. A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster. Biochem. J. 2012;442:181–190. doi: 10.1042/BJ20111747. [DOI] [PubMed] [Google Scholar]
- 14.Sayre L.M., Zelasko D.A., Harris P.L., Perry G., Salomon R.G., Smith M.A. 4-Hydroxynonenal-derived advanced lipid peroxidation end products are increased in Alzheimer's disease. J. Neurochem. 1997;68:2092–2097. doi: 10.1046/j.1471-4159.1997.68052092.x. [DOI] [PubMed] [Google Scholar]
- 15.Yoritaka A., Hattori N., Uchida K., Tanaka M., Stadtman E.R., Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 1996;93:2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Awasthi Y.C., Ansari G.A., Awasthi S. Regulation of 4-hydroxynonenal mediated signaling by glutathione S-transferases. Methods Enzymol. 2005;401:379–407. doi: 10.1016/S0076-6879(05)01024-4. [DOI] [PubMed] [Google Scholar]
- 17.Saisawang C., Ketterman A.J. Micro-plasticity of genomes as illustrated by the evolution of glutathione transferases in 12 Drosophila species. PLoS One. 2014;9:e109518. doi: 10.1371/journal.pone.0109518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wongsantichon J., Ketterman A.J. An intersubunit lock-and-key ‘Clasp’ motif in the dimer interface of delta class glutathione transferase. Biochem. J. 2006;394:135–144. doi: 10.1042/BJ20050915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winayanuwattikun P., Ketterman A.J. An electron-sharing network involved in the catalytic mechanism is functionally conserved in different glutathione transferase classes. J. Biol. Chem. 2005;280:31776–31782. doi: 10.1074/jbc.M502612200. [DOI] [PubMed] [Google Scholar]
- 20.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 2010;43:186–190. doi: 10.1107/S0021889809045701. [DOI] [Google Scholar]
- 21.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Winayanuwattikun P., Ketterman A.J. Glutamate 64, a newly identified residue of the functionally conserved electron-sharing network contributes to catalysis and structural integrity of glutathione transferases. Biochem. J. 2007;402:339–348. doi: 10.1042/BJ20061253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vararattanavech A., Ketterman A.J. A functionally conserved basic residue in glutathione transferases interacts with the glycine moiety of glutathione and is pivotal for enzyme catalysis. Biochem. J. 2007;406:247–256. doi: 10.1042/BJ20070422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding Y., Ortelli F., Rossiter L.C., Hemingway J., Ranson H. The Anopheles gambiae glutathione transferase supergene family: annotation, phylogeny and expression profiles. BMC Genomics. 2003;4:35–50. doi: 10.1186/1471-2164-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winayanuwattikun P., Ketterman A.J. Catalytic and structural contributions for glutathione binding residues in a delta class glutathione S-transferase. Biochem. J. 2004;382:751–757. doi: 10.1042/BJ20040697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubatsch I., Ridderström M., Mannervik B. Human glutathione transferase A4-4: an Alpha class enzyme with high catalytic efficiency in the conjugation of 4-hydroxynonenal and other genotoxic products of lipid peroxidation. Biochem. J. 1998;330:175–179. doi: 10.1042/bj3300175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao T., Singhal S.S., Piper J.T., Cheng J., Pandya U., Clark-Wronski J., Awasthi S., Awasthi Y.C. The role of human glutathione S-transferases hGSTA1-1 and hGSTA2-2 in protection against oxidative stress. Arch. Biochem. Biophys. 1999;367:216–224. doi: 10.1006/abbi.1999.1277. [DOI] [PubMed] [Google Scholar]
- 33.Balogh L.M., Le Trong I., Kripps K.A., Shireman L.M., Stenkamp R.E., Zhang W., Mannervik B., Atkins W.M. Substrate specificity combined with stereopromiscuity in glutathione transferase A4-4-dependent metabolism of 4-hydroxynonenal. Biochemistry. 2010;49:1541–1548. doi: 10.1021/bi902038u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranson H., Hemingway J. Mosquito glutathione transferases. Methods Enzymol. 2005;401:226–241. doi: 10.1016/S0076-6879(05)01014-1. [DOI] [PubMed] [Google Scholar]
- 35.Molin E.U., Mattsson J.G. Effect of acaricides on the activity of glutathione transferases from the parasitic mite Sarcoptes scabiei. Parasitology. 2008;135:115–123. doi: 10.1017/S0031182007003472. [DOI] [PubMed] [Google Scholar]
- 36.Lee K.W., Raisuddin S., Rhee J.S., Hwang D.S., Yu I.T., Lee Y.M., Park H.G., Lee J.S. Expression of glutathione S-transferase (GST) genes in the marine copepod Tigriopus japonicus exposed to trace metals. Aquat. Toxicol. 2008;89:158–166. doi: 10.1016/j.aquatox.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 37.Zhao D., Chen L., Qin C., Zhang H., Wu P., Zhang F. A delta-class glutathione transferase from the Chinese mitten crab Eriocheir sinensis: cDNA cloning, characterization and mRNA expression. Fish Shellfish Immunol. 2010;29:698–703. doi: 10.1016/j.fsi.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Oakeshott J.G., Johnson R.M., Berenbaum M.R., Ranson H., Cristino A.S., Claudianos C. Metabolic enzymes associated with xenobiotic and chemosensory responses in Nasonia vitripennis. Insect Mol. Biol. 2010;19(Suppl 1):147–163. doi: 10.1111/j.1365-2583.2009.00961.x. [DOI] [PubMed] [Google Scholar]
- 39.Benton M.J., Donoghue P.C. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 40.Grimaldi D., Engel M.S. Evolution of the Insects. New York: Cambridge University Press; 2005. [Google Scholar]
- 41.Wiegmann B.M., Trautwein M.D., Winkler I.S., Barr N.B., Kim J.W., Lambkin C., Bertone M.A., Cassel B.K., Bayless K.M., Heimberg A.M., et al. Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5690–5695. doi: 10.1073/pnas.1012675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engel M.S., Grimaldi D.A. New light shed on the oldest insect. Nature. 2004;427:627–630. doi: 10.1038/nature02291. [DOI] [PubMed] [Google Scholar]
- 43.Blagoderov V., Grimaldi D.A., Fraser N.C. How time flies for flies: diverse Diptera from the Triassic of Virginia and early radiation of the order. Am. Mus. Novit. 2007;3572:1–39. doi: 10.1206/0003-0082(2007)509[1:HTFFFD]2.0.CO;2. [DOI] [Google Scholar]
- 44.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]