Figure 5. Graphical representation of amino acid multiple sequence alignment for glutathione binding residues of Epsilon and Delta class GSTs from D. melanogaster and A. gambiae genomes.
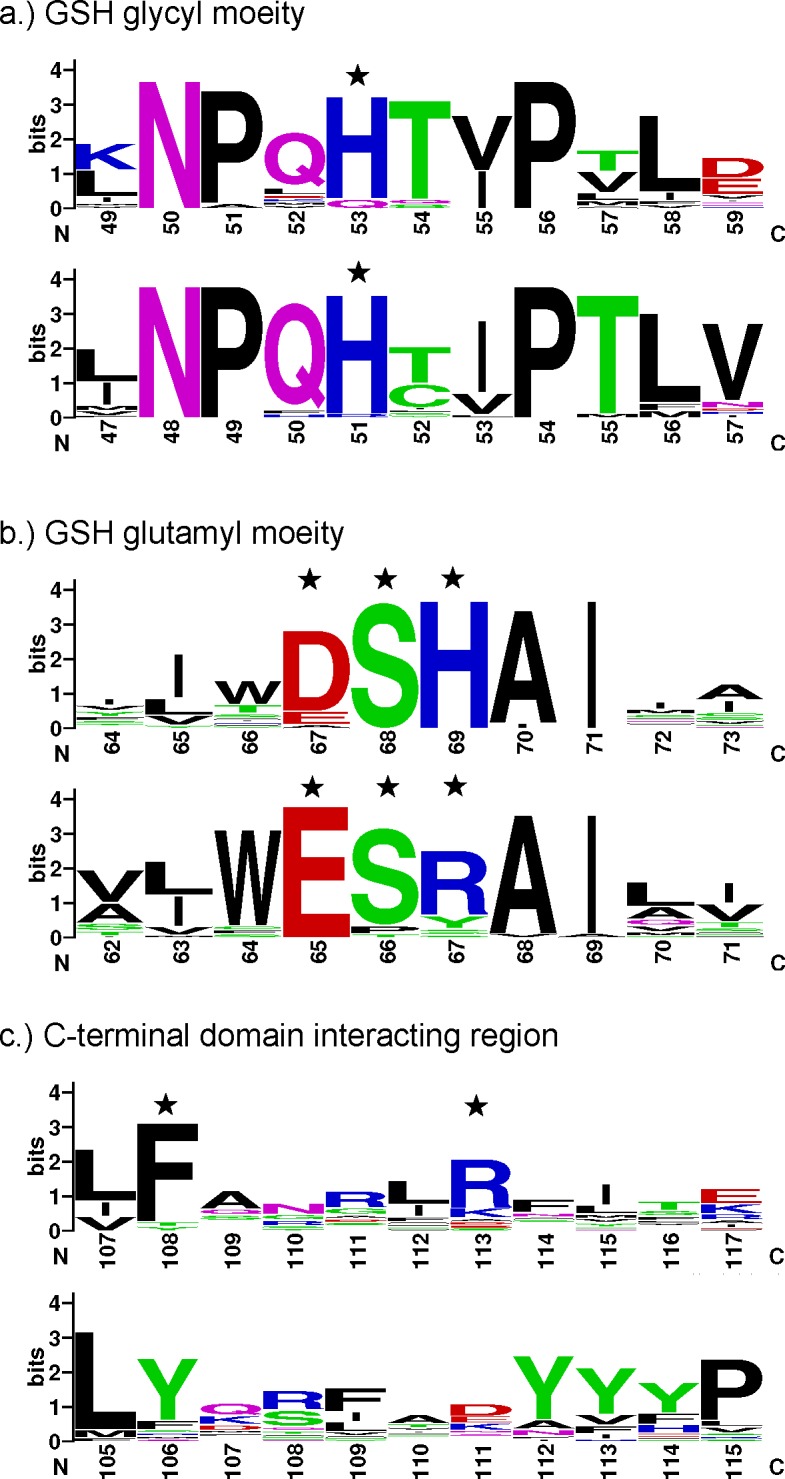
The glutathione interacting residues are indicated with stars. The top panel in each category represents Epsilon class GSTs and the bottom panel represents Delta class GSTs with respective residue numbers on an X-axis. The image was generated by WebLogo [44,45]. A logo represents each column of the alignment by a stack of letters, with the height of each letter proportional to the observed frequency of the corresponding amino acid, and the overall height of each stack proportional to the sequence conservation at that position [45]. The letters of each stack are ordered from most to least frequent, so that one may read the consensus sequence from the tops of the stacks [45]. Single letter amino acid symbols are coloured according to their chemical properties; polar amino acids (G, S, T, Y, C, Q, N) are green, basic (K, R, H) blue, acidic (D and E) red, and hydrophobic (A, V, L, I, P, W, F, M) amino acids are black [45].
