Fig. 3.
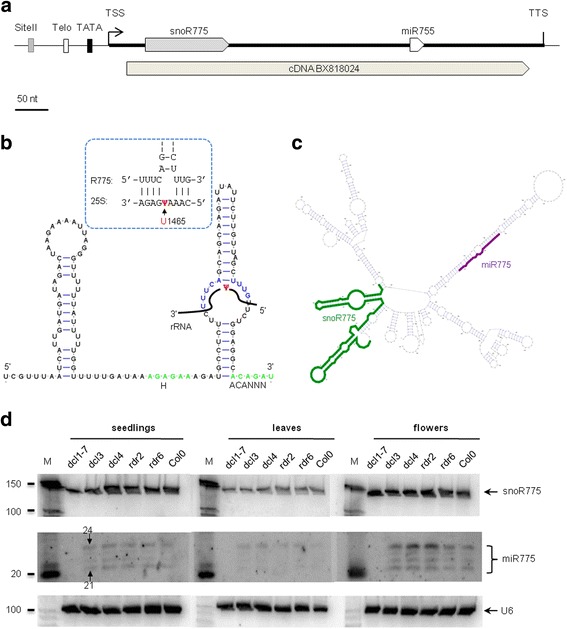
Schematic representation and experimental validation of the sno-miR775 gene. (a) Gene organization of sno-miR775. snoR775 and miR775 are denoted by a gray pentagon and an open pentagon, respectively. The TeloSII elements and TATA box are represented by different boxes: Telo-box (open box), Site II (gray box), and TATA box (black box). Both snoR775 and pre-miR775 are covered by cDNA BX818024. The transcription start site (TSS) and transcription termination site (TTS) were determined by 5’ RACE and 3’ RACE, respectively. (b) The centroid secondary structure of snoR775, drawn by using the RNAfold program. The H and ACA boxes are denoted by green color. The complementary region to the predicted target 25S rRNA is represented by blue color, and details are shown in the blue dashed box. Ψ: pseudouridylation site. (c) Secondary structure of the sno-miR775 precursor sequence. Mature miR775 and the predicted snoRNA are in purple and green colors, respectively. (d) Northern blot analysis of sno-miR775. The hybridization was carried out in 3 different tissues: 2-wk-old seedlings, 3-wk-old leaves and 5-wk-old flowers. Lanes in the blots represent the following samples: dcl1-7, dcl3, dcl4, rdr2, and rdr6as well as wild-type Col0. U6 was used as the loading control
