Fig. 4.
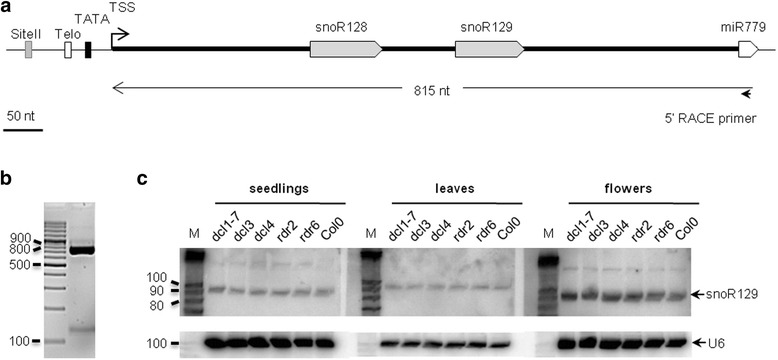
Schematic representation and experimental validation of the sno-miR779 gene. (a) Gene organization of sno-miR779. The snoRNAs and miR779 are denoted by gray pentagon and open pentagon, respectively. The TeloSII elements and TATA box are represented by different boxes: Telo-box (open box), Site II (gray box), and TATA box (black box). The transcription start site (TSS) was determined by 5’ RACE. (b) 5’ RACE amplification of snoR779. A single major signal in 5’ RACE PCR indicates the transcription start site (TSS); the product size, confirmed by sequencing, was 815 nt. (c) Northern blot analysis of snoR129. The hybridization was carried out in 3 different tissues: 2-wk-old seedlings, 3-wk-old leaves and 5-wk-old flowers. The lines in the blots include the following samples: dcl1-7, dcl3, dcl4, rdr2, and rdr6 mutants and wild type Col0. U6 was used as the loading control
