Abstract
Telomeres, which protect the ends of chromosomes, are shortened in several hematologic malignancies, often with adverse prognostic implications, but their effect on prognosis of classic and variant hairy cell leukemia (HCL and HCLv) has not been reported. HCL/HCLv genomic DNA from 46 patients was studied by PCR to determine the ratio of telomere to single copy gene number (T/S). T/S was unrelated to diagnosis of HCL or HCLv (p=0.27), but shorter T/S was associated with unmutated immunoglobulin rearrangements (p=0.033) and age above the median at diagnosis (p=0.017). Low T/S was associated with shorter overall survival from diagnosis (OS), particularly T/S <0.655 (p=0.0064, adjusted p=0.019). Shorter OS was also associated with presence of unmutated (p<0.0001) or IGHV4-34+ (p<0.0001) rearrangements, or increasing age (p=0.0002). Multivariable analysis with Cox modeling showed that short T/S along with either unmutated or IGHV4-34+ rearrangements remained associated with reduced OS (p=0.0071, p=0.0024, respectively) after age adjustment. While T/S is relatively long in HCL and the disease usually indolent with excellent survival, shortened telomeres in HCL/HCLv are associated with decreased survival. Shortened T/S could represent a risk factor needing further investigation/intervention to determine if non-chemotherapy treatment options, in addition to or instead of chemotherapy, might be particularly useful.
Keywords: Telomere, hairy cell leukemia, molecular marker, chromosomes, DNA damage
Introduction
Classic hairy cell leukemia (HCL) is a B-cell malignancy with distinctive immunophenotype, typically expressing BRAF V600E mutation, CD20, CD22, CD25, CD11c, CD103, CD123, annexin A1 (Anxa1), and tartrate-resistant acid phosphatase (TRAP) (1–4). Purine analog therapy is highly effective, with most patients achieving durable complete remissions (CR) (5, 6). HCL variant (HCLv) was first identified by Cawley et al. (7) and recently recognized by the World Health Organization as a separate disorder (4). HCLv lacks CD25, annexin A1, TRAP, and BRAF V600E, and patients respond poorly to purine analogs, with only partial response in less than 50%, CR in less than 10% and relatively poor overall survival (OS) from diagnosis (3, 8–11). We recently reported that HCL expressing the immunoglobulin heavy-chain variable (IGHV) rearrangement type IGHV4-34 expresses wild-type BRAF and has a poor prognosis like HCLv, whether immunophenotypically consistent with HCLv or classic HCL (10, 12). Mutations other than BRAF V600E have been found in HCLv and IGHV4-34+ HCL, including several within the MAPK pathway (13).
Telomerase activity (TA) prevents the further shortening of telomeres, composed of arrays of TTAGGG DNA repeats up to 25 kb, which associate with specific nucleoproteins to protect the 3’ ends of chromosomes from translocations, double-strand breaks and recombinations (14). Multiple cell divisions can result in successively shorter telomere length (TL), leading to age-related senescence including apoptosis, but high TA can be associated with immortalization and development of leukemias and lymphomas (15). Although naive normal B-cells lack TA, germinal center (GC) B-cells, which undergo at least 20 cell divisions, have high TA, thus preventing TL shortening (16).
In the B-cell malignancies, including chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), lymphoplasmacytic lymphoma (LPL), and Waldenström’s macroglobulinemia (WM), TA and TL have been observed to vary with aggressiveness and disease type (15, 17–19). In CLL, TL is highly variable and short telomeres have been associated with unmutated IGHV status, high-risk genomic aberrations, and poor outcome (15, 20, 21). In contrast to CLL, TL in MCL and DLBCL was not associated with prognosis or disease outcome (17, 18). One of these studies contained 12 cases of HCL (18), and showed that TL was longer in HCL and LPL than in FL or non-GC-like DLBCL, suggesting a stronger relationship of HCL to the GC-B-cell. TL data have not been reported for HCLv. To study telomeres in both HCL and HCLv, we determined relative TL (RTL) in patients with these B-cell disorders.
Materials and methods
Patients and blood samples
Blood was obtained from 46 patients with either HCL or HCLv as part of Institutional Review Board (IRB)-approved sample collection protocols at NCI. DNA was obtained from blood in sodium heparin tubes by the Qiagen AllPrep kit (Qiagen, Valencia, CA). HCL/HCLv cells were purified by Ficoll centrifugation followed by negative isolation of B-cells to remove T-cells, NK cells, monocytes, and other non-B-cells using the Dynabeads Untouched Human B Cells Kit (Thermo Fisher, Grand Island, NY). Samples finally had >90% HCL/HCLv cells provided that the leukemic cells prior to purification comprised >90% of the B-cells, as determined by flow cytometry. IGHV sequencing to determine closest germline sequence and % homology to germline was performed as previously described (10, 12).
Telomere Length Analysis
The monochrome multiplex quantitative PCR (MMQPCR) method (22, 23) was performed using the Rotor-Gene Corbett 3000 (Qiagen). This method has been used for many years and is reliable with low intra-individual variability (24–26). Samples chosen for DNA preparation contained >90% HCL cells, to insure that telomere data would pertain to the malignant cells and not the normal cells in the sample. The telomere and single copy gene (SCG) albumin primers were used at a final reaction concentration of 900 nM each: telg, ACACTAAGGTTTGGGTTTGGGTTTGGGTTTGGGTTAGTGT; telc, TGTTAGGTATCCCTATCCCTATCCCTATCCCTATCCCTAACA; albu, CGGCGGCGGGCGGCGCGGGCTGGGCGGaatgctgcacagaatccttg; albd, GCCCGGCCCGCCGCGCCCGTCCCGCCGgaaaagcatggtcgcctgtt. Normal female DNA dilutions for the standard curve contained a final amount of 200ng, 50ng, 12.5ng, 3.125ng in their respective tubes at a final volume of 15μl. Samples to be measured for TL and the positive control each contained a final amount of 20ng DNA in a total volume of 15μl. The standard curve was performed in duplicate for each run and samples were processed in triplicate. Syto 82 Orange Fluorescent Nucleic Acid Stain (Molecular Probes) and Quantitect Multiplex NoRox PCR Master Mix (Qiagen) were used at a final reaction concentration of 5 μM and 1X, respectively. The following were the thermal conditions: initial holding stage 1 (95°C, 15min), cycling stage 2 of two repeats (94°C, 15s; 49°C, 15s), cycling stage 3 of 28 repeats (94°C, 15s; 62°C, 10s; 74°C, acquired reading, 15s; 84°C, 10s; 88°C, acquired reading, 15s), and melt curve (ramp from 75°C to 95°C, rising by 0.2° each step, waiting for 60s on the first step and 15s for each step afterwards). The Ct values acquired at 74°C and 88°C of cycling stage 3 correspond to the telomere and albumin signals, respectively. The MMQPCR data was analyzed using the Rotor-Gene 6 software version 6.1. Two standard curves were generated for telomere and SCG albumin using the auto-find threshold option. Using each sample’s telomere Ct and albumin Ct, telomere ng and albumin ng were interpolated from their respective standard curve. Telomere ng/Albumin ng generated T/S ratios for each reaction. The average T/S ratio from samples run in triplicate and standard deviation values were used in our analysis.
Statistical methods
Dichotomous patient characteristics were compared between two groups using a Fisher’s exact test, and an exact form of the Wilcoxon rank sum test was used for comparing continuous parameters between two groups. For these analyses, the p-values are reported without adjustment for multiple comparisons since they are considered exploratory and descriptive.
The correlation between continuous parameters was determined using Spearman non-parametric correlation analysis. For these analyses, |r| >0.70 would be considered a strong correlation; 0.50 < |r| < 0.70 would be considered moderately strong; 0.30 < |r| < 0.50 would be considered weak to moderately strong, and if |r| < 0.30, the correlation is weak. Since the p-value for a correlation coefficient is a test of whether r=0, this is less important in interpreting the result than the strength of the correlation itself.
Survival was determined from date of diagnosis until date of death or date last reported to be alive. Survival probabilities as a function of time were determined by the Kaplan-Meier method, with the significance of the difference among curves compared using the log-rank test. The T/S ratio was divided into quartiles and the patients with values in these 4 categories were initially compared with respect to their association with survival. Subsequently, a single division was made to separate patients into two groups on the basis of the best association with survival. In this case, the p-value was adjusted by multiplying the unadjusted p-value by 3 to account for the implicit number of tests needed to arrive at the ultimate division. Age was also explored in a similar fashion and divided into three categories based on the survival curves. To further assess the importance of the multiple factors considered jointly for their association with survival, a Cox proportional hazards model analysis was performed.
All p-values are two-tailed, and except as noted above, are presented without any adjustment for multiple comparisons.
Results
Patients tested, clinical factors
Of the 46 patients tested, 27 had classic HCL and 19 had the variant HCLv. As shown in Table 1, median age at diagnosis was lower for HCL than for HCLv (46.6 vs 55.3 years, p=0.028). Males outnumbered females 37 to 9, with no difference in ratio between HCL and HCLv (p=0.46). The median number of prior courses of purine analog, including not only cladribine (CdA) and pentostatin (DCF), but also bendamustine and fludarabine, was 3 (range: 0–8) for HCL and one (range: 0–6 ) for HCLv (p=0.016). The higher number of purine analogs for HCL compared to HCLv was expected since the more rapid progression in the latter results in patients presenting earlier for clinical trials. Median white blood cell (WBC) count at the time of T/S measurement was 28.4 (range: 0.8–141) per nL in HCL vs 25.3 (range: 4.99–304) in HCLv (p=0.51). All patients had active disease with median 7.14 HCL cells/nL (range: 0.00092–134) vs 19.2 (range: 0.475–286) HCLv cells/nL in the blood (p=0.71), as determined by flow cytometry. Seventeen of 27 (63%) with HCL had prior splenectomy, vs 7 of 19 (37%) with HCLv (p=0.13). While WBC and HCL counts were not significantly different between HCL and HCLv, they were different as expected in patients with vs. without prior splenectomy (Table 1, p=0.020, p=0.031).
Table 1.
Patient characteristics in HCL and HCLv**
| HCL (n=27) | HCLv (n=19) | P-value | |
|---|---|---|---|
| Age at diagnosis: | 46.6 (29.6–80.1) | 55.3 (37.1–79.8) | 0.028 |
| Male/Female | 23/4 | 14/5 | 0.46 |
| Prior purine analog courses | 3 (0–8) | 1 (0–6) | 0.016 |
| WBC (/nL) | 28.4 (0.8–141) | 25.3 (4.99–304) | 0.51 |
| HCL count (/nL) | 7.14 (0.00092–134) | 19.2 (0.475–286) | 0.71 |
| Prior splenectomy | 17 of 27 | 7 of 19 | 0.13 |
| Unmutated IGHV | 5 of 27 | 12 of 19 | 0.0045 |
| IGHV4-34+ | 5 of 27 | 7 of 19 | 0.19 |
| Splenectomy (n=24) | No Splenectomy (n=22) | P-value | |
| WBC (/nL) | 46.75 (1.43–304) | 13.45 (0.8–120) | 0.020 |
| HCL count (/nL) | 2.75 (0.229–286) | 8.78 (0.00092–109) | 0.031 |
| Mutated* (n=29) | Unmutated (n=17) | P-value | |
| Age at diagnosis | 45.8 (29.6–72.9) | 56.7 (42.4–80.1) | 0.0010 |
| Male/Female | 23/6 | 14/3 | 1.0 |
| WBC (cells/nL) | 19.3 (0.8–304) | 28.5 (4.99–120) | 0.39 |
| HCL count (cells/nL) | 10.5 (0.00092–286) | 26.4 (0.475–109) | 0.54 |
| Prior splenectomy | 17 of 29 | 7 of 17 | 0.36 |
| Prior purine analog courses | 2 (0–8) | 1 (0–3) | 0.061 |
| IGHV4-34+ | 0 of 29 | 12 of 17 | <0.0001 |
Mutated IGHV is defined as ≤ 98% homology to germline sequence
Values for continuous data are expressed as medians (ranges)
Clinical factors vs homology to germline
To be able to relate telomere data to homology of the IGHV sequence relative to its closest germline sequence, homology was first compared to clinical factors. Unmutated IGHV, defined as >98% homology to germline, was observed in 5 of 27 (19%) HCL vs 12 of 19 (63%) HCLv patients (p=0.0045). Compared to patients with mutated rearrangements, patients with unmutated rearrangements had higher age at diagnosis (median 56.7 vs 45.8 years, p=0.0010), and showed a trend for fewer purine analog courses before presenting for protocol therapy (median 1 vs 2 courses, p=0.061). The 12 IGHV4-34+ HCL/HCLv patients all had unmutated rearrangements. As shown in Table 1, neither male/female ratio, WBC or HCL count, or history of prior splenectomy was significantly different between mutated and unmutated cases (p=0.36 to p=1.0).
Telomere length in HCL and HCLv
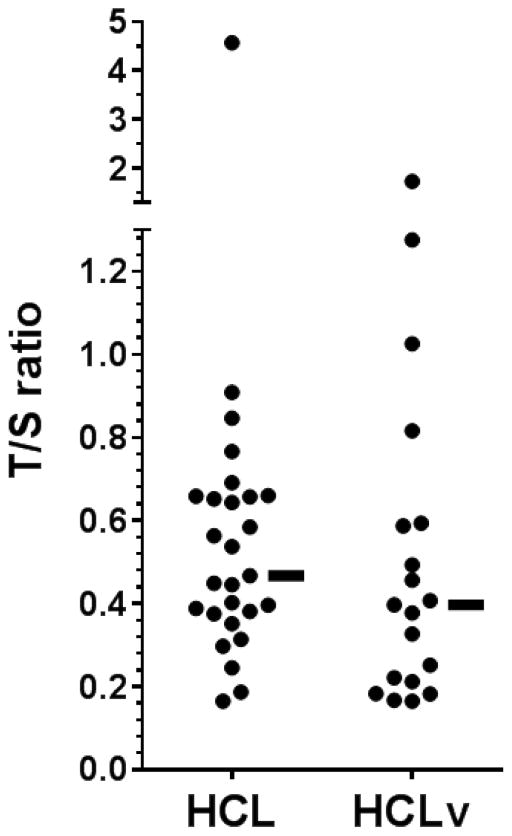
The ratio of telomere (T) copy number to single (S) copy gene number (the T/S value) was obtained for 27 HCL and 19 HCLv patients. As shown in Figure 1 and Table 2A, T/S ratio varied considerably, from 0.17 to 4.57 (median 0.47) in HCL, and 0.17–1.73 (median 0.40) in HCLv, p=0.27. Median T/S ratios were higher in patients younger vs older than 54 years (0.58 vs 0.39, p=0.017). T/S ratio did not differ with respect to sex (p=0.74) or history of prior splenectomy (p=0.58). T/S ratios were significantly higher in mutated vs unmutated cases (median 0.56 vs 0.39, p=0.033), and there was a trend (p=0.082) for higher T/S ratios in mutated cases when HCLv patients were considered separately. Finally, T/S ratio was not related to whether patients responded to initial cladribine treatment (p=1.00). Thus comparison of T/S ratios between 2 groups of patients showed at least trends for larger T/S ratios in patients who were either younger or had mutated IGHV rearrangements.
Figure 1. T/S ratios.
27 HCL and 19 HCLv patients. Median values, indicated by horizontal bars, were 0.47 for HCL and 0.40 for HCLv.
Table 2A.
comparison of T/S medians between groups
| medians | P-value | |
|---|---|---|
| HCL vs HCLv | 0.47 vs 0.40 | 0.27 |
| IGHV4-34 negative vs positive | 0.45 vs 0.40 | 0.58 |
| HCL & IGHV-34-neg vs others | 0.515 vs 0.397 | 0.23 |
| Age at diagnosis < vs > 54 | 0.58 vs 0.39 | 0.017 |
| Males vs Females | 0.45 vs 0.40 | 0.74 |
| Splenectomy or not | 0.40 vs 0.56 | 0.58 |
| Mutated vs unmutated | 0.56 vs 0.39 | 0.033 |
| Mutated vs unmutated (HCL) | 0.52 vs 0.40 | 0.52 |
| Mutated vs unmutated (HCLv) | 0.59 vs 0.31 | 0.082 |
| CdA response vs no response | 0.52 vs 0.46 | 1.00 |
Correlations of T/S ratio with clinical factors
To determine if T/S ratios were correlated with continuous clinical and laboratory variables in the 46 patients studied, Spearman non-parametric correlation analysis was performed. As shown in Table 2B, only homology of IGHV rearrangement to germline sequence and age had slightly more than weak, negative correlations with T/S ratio, with r=−0.33 and r=−0.38, respectively, and r=−0.442 for T/S ratio vs age in HCLv patients. The T/S ratio correlation with the number of prior purine analog courses, or circulating WBC or HCL cells was very weak (Table 2B).
Table 2B.
Spearman correlation analysis of T/S ratios with clinical factors
| Factor | n | r | P-value |
|---|---|---|---|
| Homology to germline | 46 | −0.331 | 0.025 |
| Homology (HCL only) | 27 | −0.055 | 0.78 |
| Homology (HCLv only) | 19 | −0.349 | 0.14 |
| Age at diagnosis | 46 | −0.379 | 0.0094 |
| Age at diagnosis (HCL only) | 27 | −0.139 | 0.49 |
| Age at diagnosis (HCLv only) | 19 | −0.442 | 0.058 |
| Prior purine analog courses | 46 | 0.068 | 0.65 |
| WBC (cells/nL) | 46 | −0.157 | 0.30 |
| HCL count (cells/nL) | 46 | −0.192 | 0.20 |
Survival with respect to clinical factors
To better understand the relationship between survival and T/S ratio in HCL and/or HCLv, we created Kaplan-Meier curves, comparing groups of patients using the log-rank test. As shown in Table 3, OS was not significantly shorter in HCLv compared to HCL (p=0.54), but both IGHV4-34+ (p<0.0001) and unmutated rearrangement (p<0.0001) were associated with a poor prognosis for survival. Age at diagnosis also was associated with lower OS. Quartiles for age at diagnosis < 42, 42–54, 54–62, and >62 had significantly different OS (global p=0.0004). Differences in survival were present between ages <42 and 42–54 (p=0.054) and between 42–54 and 54–62 (p=0.027), but not between ages 54–62 and >62 (p=0.48); thus, the upper two age groups were combined for the subsequent evaluations. Survival in the remaining 3 age groups are shown in Table 3 and Figure 2A, with global p=0.0002 and a significant difference between 42–54 and >54 (p=0.016, unadjusted). Thus, Table 3 shows that survival did not depend on diagnosis of HCL vs HCLv, but that patients survived longer if rearrangements were mutated, were negative for IGHV4-34, or if patients were younger at diagnosis.
Table 3.
Survival from diagnosis with respect to T/S and clinical factors
| Factor | Median survival (years) | P-value |
|---|---|---|
| Individual parameter results | ||
| HCL vs HCLv | 13.7 vs 8.9 | 0.54 |
| IGHV4-34 negative vs positive | 17.0 vs 8.2 | <0.0001 |
| Mutated (M) vs unmutated (U) | 17.0 vs 8.2 | <0.0001 |
| Age <42, 42–54, 54–62, >62 | 31.5, 14.6, 9.0, 5.3 | 0.0004 global |
| <42 vs 42–54 | 0.054 | |
| 42–54 vs 54–62 | 0.027 | |
| 54–62 vs >62 | 0.48 | |
| Age <42, vs 42–54 vs >54 | 31.5, 14.6, 8.9 | 0.0002 global |
| 42–54 vs >54 | 0.016 | |
| T/S <0.3, 0.3–0.447, 0.448–0.655, >0.655 | 10.8, 9.0, 8.2, not reached | 0.042 global |
| T/S < vs > 0.655 | 9.0 vs not reached | 0.0064* |
| T/S ratio along with mutation or IGHV4-34+ status | ||
| <0.655(U) vs <0.655(M) vs >0.655 | 5.3, 13.2, not reached | <0.0001 |
| <0.655(U) vs <0.655(M) | <0.0001 | |
| <0.655(M) vs >0.655 | 0.076 | |
| <0.655(U) vs >0.655 | <0.0001 | |
| <0.655(V) vs <0.655(O) vs >0.655 | 5.3, 13.2, not reached | <0.0001 |
| <0.655(V) vs <0.655(O) | 0.0006 | |
| <0.655(O) vs >0.655 | 0.041 | |
| <0.655(V) vs >0.655 | <0.0001 | |
Adjusted p-value 0.019
Abbreviations: mutated (M), unmutated (U), IGHV4-34 (V), other IGHV (O)
No patients had values equal to the above borderlines, i.e. age = 42.0, 54.0, or 62.0, or T/S 0.655.
Figure 2. Overall survival from diagnosis.
In A, according to age at diagnosis in three categories, in B, T/S ratio >0.655, or <0.655 combined with either mutated vs. un-mutated rearrangements, and in C, T/S ratio >0.655, or <0.655 combined with either IGHV4-34+ or negative rearrangements.
Survival with respect to T/S ratio
To determine if T/S ratio was related to survival, the patients were split into quartiles of 11–12 patients each with respect to T/S ratio, as shown in Table 3. Globally, T/S was related to survival (p=0.042), but the lower 3 quartiles did not significantly differ. Rather, T/S ratio (<0.655 vs. >0.655) was associated with OS (p=0.0064 unadjusted; p=0.019 adjusted). To determine whether T/S ratio retains its importance as a risk factor for survival when other factors are taken into consideration, multivariable analysis was performed. To determine whether T/S ratio influenced survival in addition to homology to germline (mutated vs unmutated), groups with T/S >0.655 or <0.655, with unmutated or mutated rearrangements, were compared. Specifically, since T/S >0.655 or <0.655, as well as mutation status of rearrangements and IGHV4-34+ status were each important factors, their combined effects on survival were explored. Since only 2 patients with T/S >0.655 had unmutated rearrangements, all patients with T/S >0.655 were considered as one group. As shown in Table 3 and Figure 2B, the three resulting homology and T/S groups demonstrated an association with survival, with global p<0.0001. There was a trend for significance between the mutated T/S <0.655 group and the T/S >0.655 group (p=0.076), despite the inclusion in the T/S >0.655 group of the 2 unmutated samples. Additionally, as shown Table 3 and Figure 2C, similar groupings based on the presence of IGHV4-34 and T/S also demonstrated a clear association with survival, with global p<0.0001. There was a trend for significance between the non-IGHV4-34 (VH-other) T/S <0.655 group and the T/S >0.655 group (p=0.041), again despite the inclusion in the T/S >0.655 group of 2 IGHV4-34+ samples.
Multivariable Cox Model
To further explore whether T/S ratio retains its status as an important risk factor for survival in HCL/HCLv, multivariable Cox Modeling was performed analyzing both age at diagnosis and T/S ratio, combined with either homology to germline (mutated or unmutated) or VH usage (IGHV4-34 or not), as found to be important in the analyses reported above. As shown in Table 4 and Figure 2, ordered homology parameters as defined were associated with progressively longer survival, while increasing age at diagnosis was associated with progressively shorter survival. The 1st Cox model shown in Table 4 demonstrates that even after adjusting for age, T/S ratio along with germline homology remained associated with survival (p=0.0071, Hazard Ratio (HR): 0.285; 95% CI for HR: 0.115–0.711). The 2nd Cox model shown in Table 4 demonstrates that after adjusting for age, T/S ratio in addition to presence of IGHV4-34 remained associated with survival (p=0.0024, HR: 0.204; 95% CI for HR: 0.073–0.569). Further modeling using all 3 factors (T/S ratio, including either presence of IGHV4-34 or homology to germline, and age at diagnosis) showed that the parameter involving IGHV4-34 lost it statistical significance in the presence of the other parameters. This was not unexpected due to the overlap of IGHV4-34+ unmutated HCL (Table 1). Thus T/S ratio was found to be significantly and positively associated with survival in HCL/HCLv, along with either germline homology or VH usage of the immunoglobulin rearrangement, independent of the age of diagnosis.
Table 4.
Multivariate Cox Model Cox proportional hazards model results.
Based on combinations of T/S ratio and homology (mutated or unmutated) or VH-type (IGHV4-34 or other), two models demonstrate the association of the ordered parameters with overall survival, adjusting for age at diagnosis.
| Model 1: | ||||
|---|---|---|---|---|
| Parameter | ||||
| Parameter | Estimate | P-value | Hazard Ratio (HR) | 95% CI for HR |
| Homology & T/S* | −1.254 | 0.0071 | 0.285 | 0.115–0.711 |
| Age at diagnosis*** | 0.9161 | 0.0172 | 2.500 | 1.177–5.310 |
| Model 2: | ||||
| Parameter | ||||
| Parameter | Estimate | P-value | Hazard Ratio (HR) | 95% CI for HR |
| VH-usage & T/S** | −1.590 | 0.0024 | 0.204 | 0.073–0.569 |
| Age at diagnosis*** | 0.842 | 0.0284 | 2.322 | 1.093–4.931 |
- Homology parameter 1: T/S ratio <0.655, unmutated
- Homology parameter 2: T/S ratio <0.655, mutated
- Homology parameter 3: T/S ratio >0.655
- VH-usage parameter 1: T/S ratio <0.655, IGHV4-34
- VH-usage parameter 2: T/S ratio <0.655, non-IGHV4-34
- VH-usage parameter 3: T/S ratio >0.655
- Age at diagnosis parameter 1: <42
- Age at diagnosis parameter 2: 42-54
- Age at diagnosis parameter 3: >54
Discussion
A normal cell undergoes senescence or apoptosis when telomere length falls below a certain threshold. In the presence of TA, telomere length is maintained and the cell evades both age-related (or cell division-related) senescence and apoptosis. Telomere maintenance may also lead to immortalization and tumorigenesis. Because telomere shortening in some hematologic disorders including CLL, but not others, has been associated with poor prognosis, we decided to study TL in HCL and HCLv. HCL and HCLv samples from 27 and 19 patients, respectively, were assayed by MMQPCR to determine ratios of relative telomere copy number to relative single copy gene number (T/S). Comparison of Kaplan-Meier survival curves did show shorter survival with T/S < 0.655 when examining the 46 patients. Moreover, while low T/S ratio was associated with unmutated rearrangements, and with more advanced age at diagnosis, and while these factors each predicted for shorter survival, multivariable analysis showed that T/S ratio retained its association with survival, particularly when combined with age and either homology or IGHV4-34 status.
Telomere dysfunction in leukemias and other hematologic disorders
Telomeres have been extensively studied in many hematologic malignancies. In the acute leukemias, it was reported that TL is shorter in ALL than in acute myelogenous leukemia (AML) at diagnosis, but the reverse was true at relapse (14, 27). Decreased TL was considered a key factor in the prognosis of acute leukemias (28). In chronic myelogenous leukemia (CML), TL was shorter in accelerated phase and blast crisis than in chronic phase, and also was shorter in CML with more rapid progression (29). In multiple myeloma (MM), TL is relatively short, and TA was associated with overall survival (30). Patients with more aggressive B-cell disorders such as Burkitt’s lymphoma have longer TL and higher TA than the more indolent tumors such as CLL (31). Ladetto et al. reported that pre-GC B-cell malignancies have shorter TL than GC-experienced tumors (19). In addition, TL was reported to be relatively short for MCL and CLL, intermediate for marginal zone lymphoma, MM and follicular lymphoma, and long for diffuse large B-cell and Burkitt’s lymphomas (19). In CLL, unmutated IGHV status with short telomeres was associated with more genetic aberrations and poorer survival than IGHV gene mutated cases (20, 32–36). Shortened TL in CLL was associated with poor outcome even in patients with early disease (21). In contrast to CLL, in both MCL and DLBCL no association was found between TL and prognosis (17, 18). In bone marrow failure syndromes, including aplastic anemia and myelodysplasia, short TL was observed and correlated to adverse prognosis (14).
Telomere activity and length in HCL
Very few studies have investigated telomeres in HCL. Trentin et al. studied TA in normal B-cells and in 35 patients with B-cell malignancies, including 13 with HCL and 2 with HCLv, and reported that like MCL and CLL, TA was significantly higher in HCL than in normal B-cells. Among the 13 with HCL, 6 with stable disease had lower TA compared to 7 with progressive disease (37). The 2 patients with HCLv also had TA levels higher than any of 7 normal B-cell samples. This study did not report TL. Walsh et al. studied TL in hematologic malignancies including 12 patients with HCL and reported that the median T/S ratio in HCL, 0.62, was greater than median T/S values of CLL, MCL, DLBCL, and FL, which were 0.46, 0.47, 0.53 and 0.53, respectively (18). The 12 HCL patients in that study were too few for statistical significance. In our study, the median T/S ratio of our 22 patients with classic IGHV4-34-negative HCL was 0.515, slightly higher than the 0.46 value previously reported for CLL, and slightly lower than the 0.62 value reported for HCL (18). It is possible that the median T/S in our study for classic HCL patients was relatively low because our patients were older or more heavily pretreated; only 3 of our classic HCL patients were previously untreated at the time HCL samples were collected. Differences in methodology might also account for the reduction in T/S. The need for samples containing >90% HCL cells likely created a selection bias for our study since this would favor more poor-prognosis patients with HCLv, IGHV4-34+ HCL, and splenectomized HCL. However, these were the prognostic factors analyzed. Interestingly, of our 4 untreated patients, one with T/S 0.816 had IGHV4-34+ HCLv, and one with T/S 0.445 had classic HCL and splenectomy due to ruptured spleen associated with a contact sport. The other 2 untreated patients, both with classic HCL and T/S values 0.643–0.584, had HCL counts 1.1–33/nl, but no high-risk factors. Of 83 patients with newly diagnosed classic HCL, 39% had HCL cells comprising >90% of normal B-cells by flow cytometry. Thus, about 40% of patients with newly diagnosed HCL would be expected to have >90% purified HCL cells after Ficoll centrifugation and negative B-cell selection, and hence evaluable as in this study for T/S analysis. Clearly, additional untreated classic HCL patients should be studied to better understand the role of TL in newly diagnosed HCL, but survival data on such patients may take several decades of follow-up.
Significance of TL in HCL and variants
The presence of many overlapping poor-prognosis groups within the 46 patients examined, including HCLv, unmutated HCL, and IGHV4-34+ HCL, gave us the opportunity to study the relationship of T/S to poor prognosis factors in HCL/HCLv. Our study shows for the first time in HCL and HCLv that low T/S is associated with advance age at diagnosis or unmutated rearrangements, and these factors, in addition to presence of IGHV4-34+ rearrangements, were associated with shorter OS. IGHV4-34+ rearrangements in HCL and HCLv have been shown to be associated with poor prognosis (12). Moreover, we found by multivariable analysis that low T/S, specifically <0.655, especially when combined with either IGHV4-34+ or unmutated rearrangements, was independently associated with shorter OS. This is consistent with the hypothesis that the telomere protects the chromosome from genetic instability (38). Indeed, patients with IGHV4-34+ or unmutated HCL or HCLv but less often classic HCL have been reported to have chromosomal aberrations which are associated with poor prognosis (8, 39–42). It may be useful to follow patients after obtaining T/S ratio at diagnosis to determine prognosis, including response to 1st line purine analog treatment. Doing so could determine if more extensive treatment, for example, rituximab combined with cladribine (8, 43), should be used in patients with short T/S but otherwise classic-appearing HCL. Finally, it might be useful to determine the relative roles of chemotherapy and non-chemotherapy treatments for HCL in hastening the shortening of telomeres. We anticipate that such studies might show further benefits for new non-chemotherapy approaches now under clinical investigation, including BTK or BRAF/MEK inhibition, or targeted immunotoxin therapy (44, 45).
Highlights.
Telomeres protect the ends of chromosomes but are sometimes shortened in leukemias.
Short telomeres in hairy cell leukemia are associated with decreased overall survival.
Short telomere length correlates with other poor prognosis factors in hairy cell leukemia.
Low telomere length should be studied further in this disease.
Acknowledgments
The authors would like to recognize the contributions of our clinical staff for helping to obtain samples during the time they were collected, including Betty Maestri, Rita Mincemoyer, Barbara Debrah and Sonya Duke. NIH summer interns Paul Tran and Gabriella Canales contributed to the initial development of the MMQPCR TL assay at NCI. This study was supported in part by the Intramural Research Program, NCI, and the Hairy Cell Leukemia Foundation.
Footnotes
Author contributions: DCE, EA, PSM and RJK designed the research study, EA, HZ, AG, and DP performed the research, EA, DCE, SMS, YW, PSM and RJK analyzed the data, and EA, DCE, SMS and RJK wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli MP, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falini B, Tiacci E, Liso A, Basso K, Sabattini E, Pacini R, et al. Simple diagnostic assay for hairy cell leukaemia by immunocytochemical detection of annexin A1 (ANXA1) Lancet. 2004;363:1869–1870. doi: 10.1016/S0140-6736(04)16356-3. [DOI] [PubMed] [Google Scholar]
- 3.Matutes E. Immunophenotyping and differential diagnosis of hairy cell leukemia. Hematol Oncol Clin North Am. 2006;20:1051. doi: 10.1016/j.hoc.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4. Vol. 2. World Health Organization; 2008. [Google Scholar]
- 5.Goodman GR, Burian C, Koziol J, Saven A. Extended follow-up of patients with hairy cell leukemia after treatment with cladribine. J Clin Oncol. 2003;21:891–896. doi: 10.1200/JCO.2003.05.093. [DOI] [PubMed] [Google Scholar]
- 6.Else M, Dearden CE, Matutes E, Garcia-Talavera J, Rohatiner AZ, Johnson SA, et al. Long-term follow-up of 233 patients with hairy cell leukaemia, treated initially with pentostatin or cladribine, at a median of 16 years from diagnosis. Br J Haematol. 2009;145:733–740. doi: 10.1111/j.1365-2141.2009.07668.x. [DOI] [PubMed] [Google Scholar]
- 7.Cawley JC, Burns G, Hayhoe FG. A chronic lymphoproliferative disorder with distinctive features: a distinct variant of hairy-cell leukaemia. Leuk Res. 1980;4:547–559. doi: 10.1016/0145-2126(80)90066-1. [DOI] [PubMed] [Google Scholar]
- 8.Kreitman RJ, Wilson W, Calvo KR, Arons E, Roth L, Sapolsky J, et al. Cladribine with immediate rituximab for the treatment of patients with variant hairy cell leukemia. Clin Cancer Res. 2013;19:6873–6881. doi: 10.1158/1078-0432.CCR-13-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiacci E, Schiavoni G, Forconi F, Santi A, Trentin L, Ambrosetti A, et al. Simple genetic diagnosis of hairy cell leukemia by sensitive detection of the BRAF-V600E mutation. Blood. 2012;119:192–195. doi: 10.1182/blood-2011-08-371179. [DOI] [PubMed] [Google Scholar]
- 10.Xi L, Arons E, Navarro W, Calvo KR, Stetler-Stevenson M, Raffeld M, et al. Both variant and IGHV4-34-expressing hairy cell leukemia lack the BRAF V600E mutation. Blood. 2012;119:3330–3332. doi: 10.1182/blood-2011-09-379339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robak T. Hairy-cell leukemia variant: recent view on diagnosis, biology and treatment. Cancer Treat Rev. 2011;37:3–10. doi: 10.1016/j.ctrv.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Arons E, Suntum T, Stetler-Stevenson M, Kreitman RJ. VH4-34+ hairy cell leukemia, a new variant with poor prognosis despite standard therapy. Blood. 2009;114:4687–4695. doi: 10.1182/blood-2009-01-201731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterfall JJ, Arons E, Walker RL, Pineda M, Roth L, Killian JK, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet. 2014;46:8–10. doi: 10.1038/ng.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones CH, Pepper C, Baird DM. Telomere dysfunction and its role in haematological cancer. Br J Haematol. 2012;156:573–587. doi: 10.1111/j.1365-2141.2011.09022.x. [DOI] [PubMed] [Google Scholar]
- 15.Davison GM. Telomeres and telomerase in leukaemia and lymphoma. Transfus Apher Sci. 2007;37:43–47. doi: 10.1016/j.transci.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Weng NP, Granger L, Hodes RJ. Telomere lengthening and telomerase activation during human B cell differentiation. Proc Natl Acad Sci U S A. 1997;94:10827–10832. doi: 10.1073/pnas.94.20.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jebaraj BM, Kienle D, Lechel A, Mertens D, Heuberger M, Ott G, et al. Telomere length in mantle cell lymphoma. Blood. 2013;121:1184–1187. doi: 10.1182/blood-2012-08-452649. [DOI] [PubMed] [Google Scholar]
- 18.Walsh SH, Grabowski P, Berglund M, Thunberg U, Thorselius M, Tobin G, et al. Telomere length and correlation with histopathogenesis in B-cell leukemias/lymphomas. Eur J Haematol. 2007;78:283–289. doi: 10.1111/j.1600-0609.2007.00817.x. [DOI] [PubMed] [Google Scholar]
- 19.Ladetto M, Compagno M, Ricca I, Pagano M, Rocci A, Astolfi M, et al. Telomere length correlates with histopathogenesis according to the germinal center in mature B-cell lymphoproliferative disorders. Blood. 2004;103:4644–4649. doi: 10.1182/blood-2003-12-4412. [DOI] [PubMed] [Google Scholar]
- 20.Veronese L, Tournilhac O, Callanan M, Prie N, Kwiatkowski F, Combes P, et al. Telomeres and chromosomal instability in chronic lymphocytic leukemia. Leukemia. 2013;27:490–493. doi: 10.1038/leu.2012.194. [DOI] [PubMed] [Google Scholar]
- 21.Lin TT, Norris K, Heppel NH, Pratt G, Allan JM, Allsup DJ, et al. Telomere dysfunction accurately predicts clinical outcome in chronic lymphocytic leukaemia, even in patients with early stage disease. Br J Haematol. 2014;167:214–223. doi: 10.1111/bjh.13023. [DOI] [PubMed] [Google Scholar]
- 22.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui Y, Cai Q, Qu S, Chow WH, Wen W, Xiang YB, et al. Association of leukocyte telomere length with colorectal cancer risk: nested case-control findings from the Shanghai Women's Health Study. Cancer Epidemiol Biomarkers Prev. 2012;21:1807–1813. doi: 10.1158/1055-9965.EPI-12-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S, Sandler DP, Carswell G, Weinberg CR, Taylor JA. Reliability and short-term intra-individual variability of telomere length measurement using monochrome multiplexing quantitative PCR. PLoS One. 2011;6:e25774. doi: 10.1371/journal.pone.0025774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siraj AK, Ozbek U, Sazawal S, Sirma S, Timson G, Al-Nasser A, et al. Preclinical validation of a monochrome real-time multiplex assay for translocations in childhood acute lymphoblastic leukemia. Clin Cancer Res. 2002;8:3832–3840. [PubMed] [Google Scholar]
- 27.Capraro V, Zane L, Poncet D, Perol D, Galia P, Preudhomme C, et al. Telomere deregulations possess cytogenetic, phenotype, and prognostic specificities in acute leukemias. Exp Hematol. 2011;39:195–202. e192. doi: 10.1016/j.exphem.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Aalbers AM, Calado RT, Young NS, Zwaan CM, Wu C, Kajigaya S, et al. Telomere length and telomerase complex mutations in pediatric acute myeloid leukemia. Leukemia. 2013;27:1786–1789. doi: 10.1038/leu.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brummendorf TH, Holyoake TL, Rufer N, Barnett MJ, Schulzer M, Eaves CJ, et al. Prognostic implications of differences in telomere length between normal and malignant cells from patients with chronic myeloid leukemia measured by flow cytometry. Blood. 2000;95:1883–1890. [PubMed] [Google Scholar]
- 30.Wu KD, Orme LM, Shaughnessy J, Jr, Jacobson J, Barlogie B, Moore MA. Telomerase and telomere length in multiple myeloma: correlations with disease heterogeneity, cytogenetic status, and overall survival. Blood. 2003;101:4982–4989. doi: 10.1182/blood-2002-11-3451. [DOI] [PubMed] [Google Scholar]
- 31.Remes K, Norrback KF, Rosenquist R, Mehle C, Lindh J, Roos G. Telomere length and telomerase activity in malignant lymphomas at diagnosis and relapse. Br J Cancer. 2000;82:601–607. doi: 10.1054/bjoc.1999.0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karhu R, Tobin G, Thunberg U, Vilpo L, Sundstrom C, Knuutila S, et al. More extensive genetic alterations in unmutated than in hypermutated cases of chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2003;37:417–420. doi: 10.1002/gcc.10227. [DOI] [PubMed] [Google Scholar]
- 33.Grabowski P, Hultdin M, Karlsson K, Tobin G, Aleskog A, Thunberg U, et al. Telomere length as a prognostic parameter in chronic lymphocytic leukemia with special reference to VH gene mutation status. Blood. 2005;105:4807–4812. doi: 10.1182/blood-2004-11-4394. [DOI] [PubMed] [Google Scholar]
- 34.Bechter OE, Eisterer W, Pall G, Hilbe W, Kuhr T, Thaler J. Telomere length and telomerase activity predict survival in patients with B cell chronic lymphocytic leukemia. Cancer Res. 1998;58:4918–4922. [PubMed] [Google Scholar]
- 35.Damle RN, Batliwalla FM, Ghiotto F, Valetto A, Albesiano E, Sison C, et al. Telomere length and telomerase activity delineate distinctive replicative features of the B-CLL subgroups defined by immunoglobulin V gene mutations. Blood. 2004;103:375–382. doi: 10.1182/blood-2003-04-1345. [DOI] [PubMed] [Google Scholar]
- 36.Roos G, Krober A, Grabowski P, Kienle D, Buhler A, Dohner H, et al. Short telomeres are associated with genetic complexity, high-risk genomic aberrations, and short survival in chronic lymphocytic leukemia. Blood. 2008;111:2246–2252. doi: 10.1182/blood-2007-05-092759. [DOI] [PubMed] [Google Scholar]
- 37.Trentin L, Ballon G, Ometto L, Perin A, Basso U, Chieco-Bianchi L, et al. Telomerase activity in chronic lymphoproliferative disorders of B-cell lineage. Br J Haematol. 1999;106:662–668. doi: 10.1046/j.1365-2141.1999.01620.x. [DOI] [PubMed] [Google Scholar]
- 38.Hackett JA, Greider CW. Balancing instability: dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene. 2002;21:619–626. doi: 10.1038/sj.onc.1205061. [DOI] [PubMed] [Google Scholar]
- 39.Hockley SL, Morgan GJ, Leone PE, Walker BA, Morilla A, Else M, et al. High-resolution genomic profiling in hairy cell leukemia-variant compared with typical hairy cell leukemia. Leukemia. 2011;25:1189–1192. doi: 10.1038/leu.2011.47. [DOI] [PubMed] [Google Scholar]
- 40.Brito-Babapulle V, Matutes E, Oscier D, Mould S, Catovsky D. Chromosome abnormalities in hairy cell leukaemia variant. Genes Chromosomes Cancer. 1994;10:197–202. doi: 10.1002/gcc.2870100308. [DOI] [PubMed] [Google Scholar]
- 41.Matutes E, Wotherspoon A, Catovsky D. The variant form of hairy-cell leukaemia. Best Pract Res Clin Haematol. 2003;16:41–56. doi: 10.1016/s1521-6926(02)00086-5. [DOI] [PubMed] [Google Scholar]
- 42.Forconi F, Sozzi E, Cencini E, Zaja F, Intermesoli T, Stelitano C, et al. Hairy cell leukemias with unmutated IGHV genes define the minor subset refractory to single-agent cladribine and with more aggressive behavior. Blood. 2009;114:4696–4702. doi: 10.1182/blood-2009-03-212449. [DOI] [PubMed] [Google Scholar]
- 43.Ravandi F, O'Brien S, Jorgensen J, Pierce S, Faderl S, Ferrajoli A, et al. Phase 2 study of cladribine followed by rituximab in patients with hairy cell leukemia. Blood. 2011;118:3818–3823. doi: 10.1182/blood-2011-04-351502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreitman RJ, Tallman MS, Robak T, Coutre S, Wilson WH, Stetler-Stevenson M, et al. Phase I trial of anti-CD22 recombinant immunotoxin moxetumomab pasudotox (CAT-8015 or HA22) in patients with hairy cell leukemia. J Clin Oncol. 2012;30:1822–1828. doi: 10.1200/JCO.2011.38.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kreitman RJ. Immunoconjugates and new molecular targets in hairy cell leukemia. Hematology Am Soc Hematol Educ Program. 2012;2012:660–666. doi: 10.1182/asheducation-2012.1.660. [DOI] [PMC free article] [PubMed] [Google Scholar]