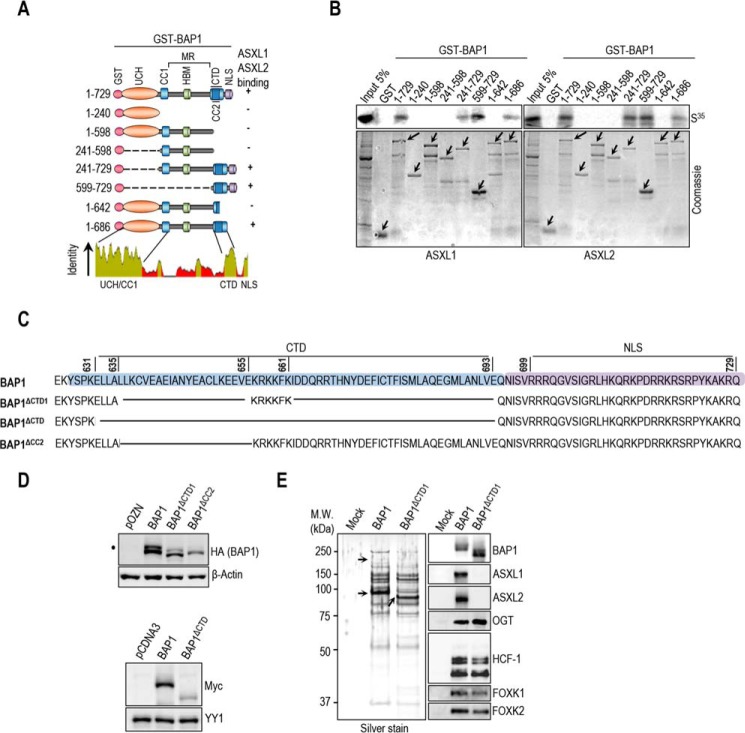
FIGURE 4.
BAP1 interacts with ASXL1/2 via its CTD domain. A, schematic representation of the BAP1 fragments used for in vitro pulldown in B. B, GST-pulldown assay using GST-BAP1 fragments and methionine 35S-labeled ASXM domains of ASXL1 or ASXL2. The arrows indicate the full-length forms of the fragments. C, schema of the different deletions in the CTD domain used to generate BAP1 mutants. BAP1ΔCTD1 represents a deletion of the CTD from 635 up to 693 amino acids except the KRKKFK motif, which is suggested to function as an NLS (21). We also generated a BAP1ΔCTD, which represents a mutant with a deletion of the CTD domain (Δ631–693 amino acids). BAP1ΔCC2 represents a mutant with a smaller deletion within the CTD domain (Δ635–655 amino acids). D, functional CTD is required for proper protein stability of BAP1. Protein expression levels of BAP1 and its CTD deletion mutant form in stable HeLa S cell lines are shown (top panel). Myc-BAP1, Myc-BAP1 ΔCTD expression constructs (3 μg each) were transfected in 293T cells, which were harvested, 3 days post-transfection, for immunoblotting (bottom panel). E, left panel, silver stain of the immunopurified BAP1 and BAP1ΔCTD1 complexes. Right panel, Western blot detection of components of the BAP1 complexes. The high and low arrows indicate the position of ASXL2 and BAP1 (WT and BAP1ΔCTD1), respectively. β-Actin or YY1 are used as protein loading controls. The dot indicates a monoubiquitinated form of BAP1 (D) (31).