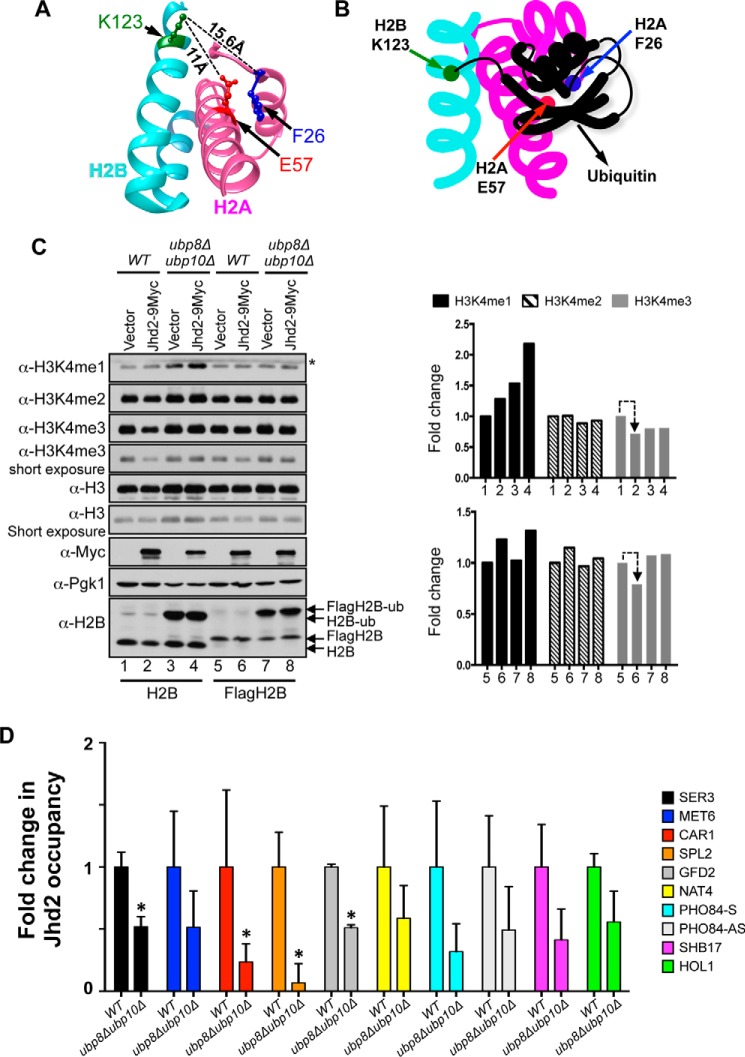
FIGURE 8.
Ubp8 and Upb10, the H2B deubiquitinases, control chromatin association and H3K4 demethylase functions of Jhd2. A, structures of H2A (magenta) and H2B (cyan) in the yeast nucleosome are shown together with the H2B Lys-123 residue (green), the site ubiquitination, and the H2A Phe-26 (blue) and Glu-57 (red) residues, the binding sites for Jhd2. Distances between H2B Lys-123 and the two H2A residues calculated using the UCSF Chimera software are shown. B, a schematic model depicting the putative masking of the H2A Phe-26 and Glu-57 residues by H2BK123 ubiquitination. C, Western blots for H3K4 methylation, H2B ubiquitination and Jhd2–9Myc were examined in control wild-type strain or in strains lacking the two H2B deubiquitinases (ubp8Δubp10Δ) and transformed with an empty plasmid vector (pRS426) or a plasmid to overexpress Jhd2–9Myc. Wild-type and ubp8Δubp10Δ in two independent yeast backgrounds, namely FY120 or Y131 (labeled as H2B and FlagH2B, respectively), were used. Crude nuclear extracts were used for examining H3K4 methylation and Jhd2 levels. Pgk1 and H3 levels serve as loading controls. For H2B ubiquitination, cells were lysed by boiling in Laemmli sample buffer. *, a cross-reacting protein. D, Jhd2-12V5 occupancies at the indicated target genes in control wild-type and ubp8Δubp10Δ strains were assessed with ChIP assays using α-V5 antibody. Following ChIP and quantitative PCR, the ChIP signal (V5 immunoprecipitation/input value) obtained from a strain without V5-tagged Jhd2 (background) was subtracted from the ChIP signal obtained from strains expressing Jhd2-12V5, and the resulting difference was defined as Jhd2 occupancy. The graph shows -fold change in Jhd2 occupancy at a given target locus in the ubp8Δubp10Δ mutant relative to Jhd2 occupancy in the control wild-type strain (set as 1). Error bars, S.E. obtained from three independent ChIP experiments. Statistical significance was calculated using Student's t test. *, p < 0.05.