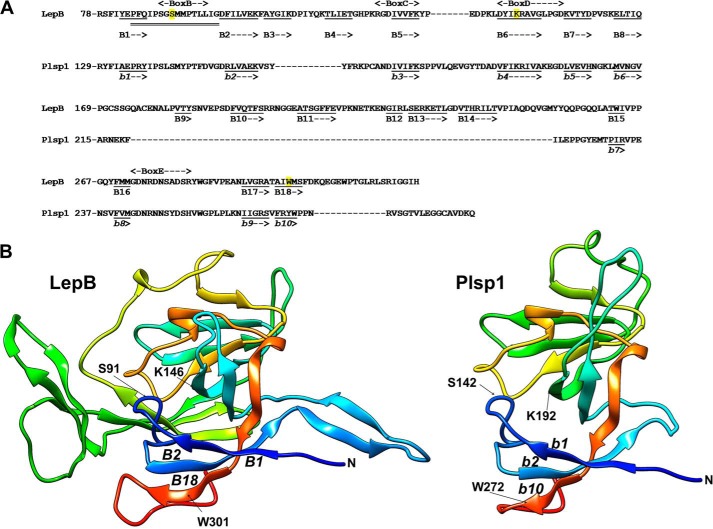
FIGURE 1.
Predicted structure of the lumenal portion of Plsp1. A, sequence alignment with the C-terminal periplasmic portion of LepB and the secondary structure prediction using Phyre2 (71) based on d1b12a (E. coli LepB; Protein Data Bank code 1b12) (5), which shows 41% sequence identity and 100% confidence for Plsp1. Shown in the diagram are the number of the most N-terminal residue in each line, conserved boxes B–E based on the work of Paetzel (2), residues in β-strands as underlined (B1–B18 for LepB and b1 to b10 for Plsp1), and the region (from Pro-84 to Gly-99) of LepB necessary for the TMD-independent membrane insertion with double line (=) (6). For LepB, the catalytic nucleophile Ser-91 and the general base Lys-146, as well as Trp-301, the residue predicted to help facilitate membrane insertion of the catalytic region (2), are highlighted in yellow. Note that the assignments to β-strands are based on the prediction and do not completely match with the ones show by Paetzel (2) and that not only the β-strands but also the region needed for membrane insertion is conserved in Plsp1. B, three-dimensional structural models of LepB78–323 (left panel) and Plsp1129–280 (right panel). The models were visualized with color by rainbow N to C terminus with the UCSF Chimera package (72), which is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS, National Institutes of Health Grant P41-GM103311). N indicates the N terminus of each structure. Indicated in LepB are the catalytic nucleophile Ser-91 (S91) and the general base Lys-146 (K146), as well as the three β-strands (B1, B2, and B18) including the essential Trp-301 (W301) (73). The corresponding catalytic residues, β-strands, and Trp in Plsp1 are indicated as S142, K192, b1, b2, b10, and W272, respectively.