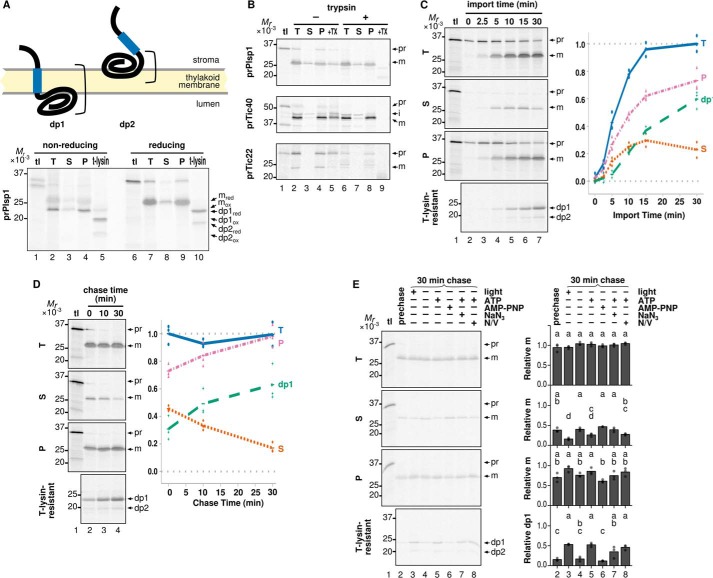
FIGURE 3.
Distribution of Plsp1 into soluble and membrane fractions after import into isolated chloroplasts. A, membrane topologies of imported Plsp1. Blue boxes denote the region corresponding to TMD (residues 111–128), and brackets indicate the portion protected from thermolysin. Radiolabeled prPlsp1 was imported into isolated chloroplasts and separated into total (T), soluble (S), pellet (P), and thermolysin-resistant (t-lysin) fractions as described in the legend to Fig. 2A. Samples were resuspended in sample loading buffer contained either no reducing agents (nonreducing) or 100 mm β-mercaptoethanol (reducing) and separated by SDS-PAGE, and protein signals were visualized using phosphorimaging. Reduced mPlsp1 (mred; detected in lanes 7–9), oxidized mPlsp1(mox; detected in lanes 2–4), reduced dp1 and dp2 (dp1red and dp2red; detected in lane 10), and oxidized dp1 and dp2 (dp1ox and dp2ox; detected in lane 5) are indicated to the right. B, in vitro chloroplast import followed by trypsin treatment and fractionation. After import of radiolabeled proteins indicated at left with 3 mm ATP in the light for 30 min at room temperature, chloroplasts were reisolated and incubated with buffer without (−) or with (+) trypsin at room temperature in the dark for 30 min. After the protease activity was quenched with trypsin inhibitor, chloroplasts were reisolated and separated into total (T), soluble (S), and pellet (P) fractions and analyzed as described above. Each lane contained samples equivalent to 3 μg of chlorophyll. As a control for trypsin activity, chloroplast samples were solubilized with 1% Triton X-100 before treatment (+TX). Bands corresponding to precursor (pr), intermediate (i; for Tic40 only), and mature (m) forms of each protein are indicated. C, import time course of Plsp1. Radiolabeled prPlsp1 was imported into isolated chloroplasts in the light with 3 mm ATP for the times indicated, followed by fractionation into total (T), soluble (S), and pellet (P) fractions and analyzed as with A. An aliquot of the total chloroplasts recovered after each time point was also lysed and incubated with thermolysin to yield dp1 and dp2. Shown at right is quantification of recovered mPlsp1 and dp1 in each fraction. The results of three repetitions and their means are shown as dots and lines, respectively. D, import-chase assay of Plsp1. Radiolabeled prPlsp1 was imported into isolated chloroplasts for 10 min in the light with 3 mm ATP, followed by treatment with 0.5 mg/ml thermolysin for 30 min on ice in the dark to remove unintegrated Plsp1 and quenching with 20 mm EDTA. Intact chloroplasts were then reisolated, incubated under light with 3 mm ATP for the times indicated, and then analyzed as described for A. Note that the EDTA treatment followed by wash with import buffer did not affect membrane insertion of Plsp1 (data not shown). E, energy requirement of membrane integration of soluble Plsp1 after import into isolated chloroplasts. Import-chase was performed and analyzed as in A under the conditions varying in the light, ATP (3 mm), AMP-PNP (3 mm), NaN3 (10 mm), and uncouplers (N/V, 1.5 μm each of nigericin and valinomycin). tl lanes were loaded as in Fig. 2. Prechase lane was loaded with the sample before the chase. Bands corresponding to prPlsp1 (pr), mPlsp1 (m), and the thermolysin degradation products (dp1 and dp2) are indicated at right. Quantification of three repetitions (circles) and their means (bars) is shown for each treatment. Means of treatments with the same letter do not differ significantly based on HSD for α = 0.05.