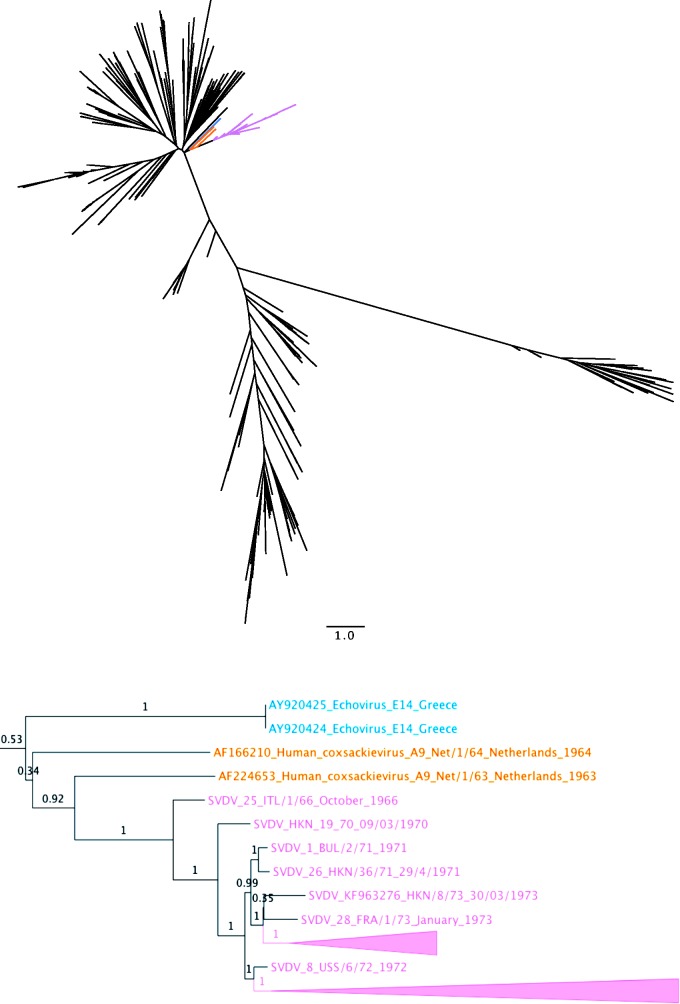
Figure 1.
Maximum likelihood tree for the 3D (Polymerase) genomic section
Result from the maximum likelihood phylogenetic analysis of the 3D (Polymerase) genomic section (see also Supplementary Table S5 and Methodology). Showing—on top—an unrooted tree with no taxa designations, except that branches leading to an SVDV isolate are coloured pink, and closest related samples are shown in orange (Dutch CV-A9 samples) and blue (Greek echovirus serotype E14 samples), giving an immediate overview of the relation between SVDV and all other sequences in the analysis, including relative (and if conferring with the 0.1 substitution-per-site bar, also absolute) distance to the nearest neighbours. Below, a cut-out of a mid-point rooted versions of the same tree. The earliest SVDV isolate (ITL/1/66) is seen as sister to all other SVDV isolates at the root of the monophyletic SVDV cluster. The early Dutch, 1963, CV-A9, isolate (Net/1/63 acc. AF224653) is seen as a strongly supported sister of all SVDV sequences (0.92 aBayes [35])