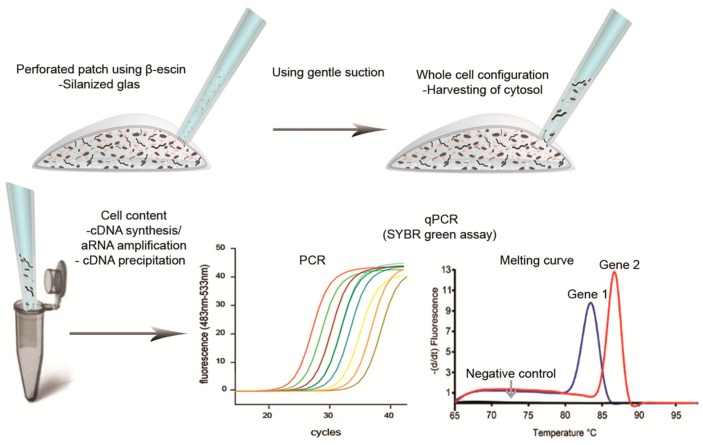
Figure 6.
Schematic overview of the protocol used in our laboratory when performing perforated patch-clamp experiments followed by single-cell qPCR. This procedure allows for using perforated patch-clamp recordings and minimizes the incidents of false positive results during gene analysis.The patch pipette is silanized by briefly exposing the tip of the glass in a 1/10 dilution of Sigmacote. To allow for a tight high-resistance interaction between the glass and cell membrane, called a gigaohm seal, the glass tip needs to be fire polished using a microforge. (Upper left) The pipette is filled with an RNase-free solution suitable for the experiments and using β-escin to perforate the cell membrane; (Upper right) the cell cytosol can be harvested following transition to whole cell configuration using gentle suction; (Lower left) the cell content is transferred to a 0.5 mL tube containing RNA stabilizing solution; (Lower middle figure) the target genes are amplified using qPCR (colored curve represents target gene been amplified); (Lower right) following qPCR amplification a melting curve analysis is performed by gently heating the PCR product(s) from 65 to 98 °C while continually reading of the fluorescent (The curve(s) are plotted as the negative 1. derivative of fluorescent with respect to temperature).