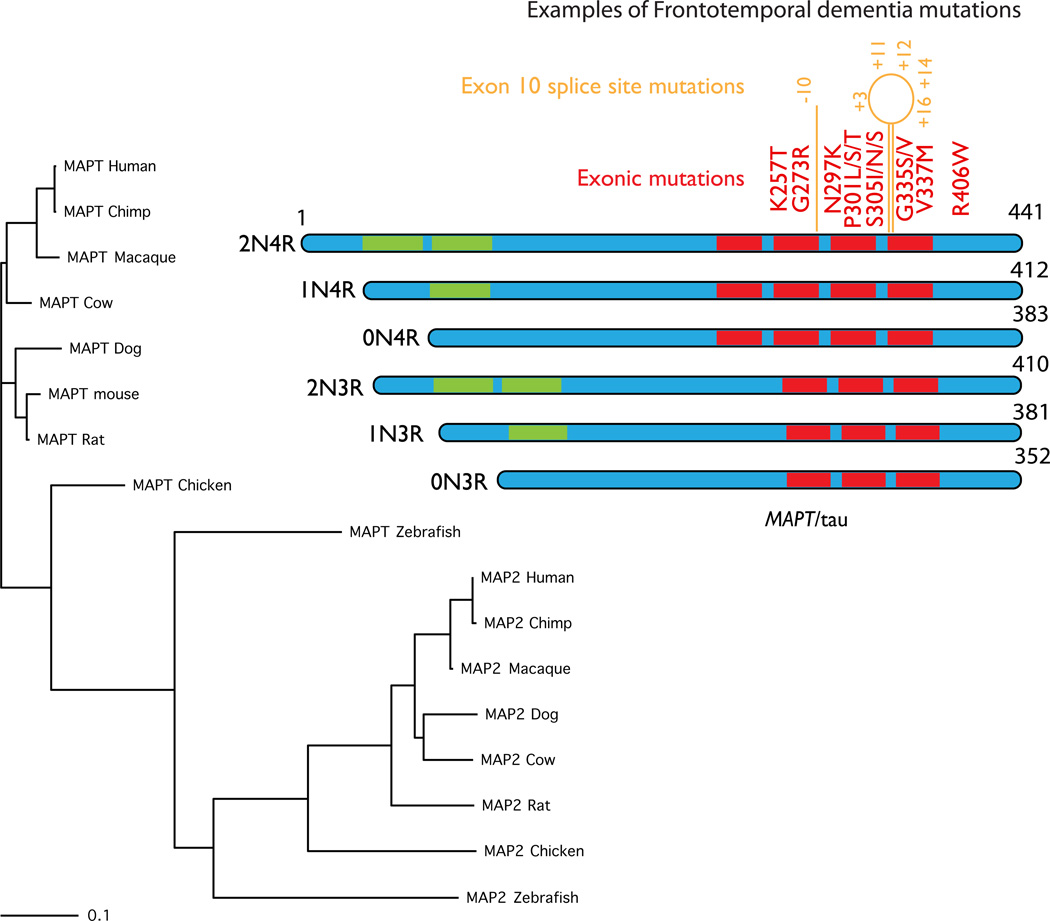
Figure 4. MAPT.
MAPT, coding for the axonal microtubule binding protein tau, is found in most vertebrates and conservation generally follows the standard phylogenetic tree for these organisms, as shown on the left of this figure. There are many other microtubule binding proteins but for clarity, here I have shown the nearest group, which includes the dendritic microtubule binding protein MAP2. These are a separate group of proteins but again these follow the same phylogenetic pattern across species. The protein structure of the six known tau isoforms is shown on the left. These isoforms are generated by alternate splicing of exons 2 and 3 (in green in the ideograms) generating different numbers of amino terminal inserts (2N, 1N or 0N) and by splicing of exon 10, which generates one extra microtubule binding repeat (4R or 3R). A selection of the known exonic FTD mutations is shown in red. In orange are intronic mutations that change the splicing but not the amino acid sequence, numbered by nucleotide position relative to exon 10.