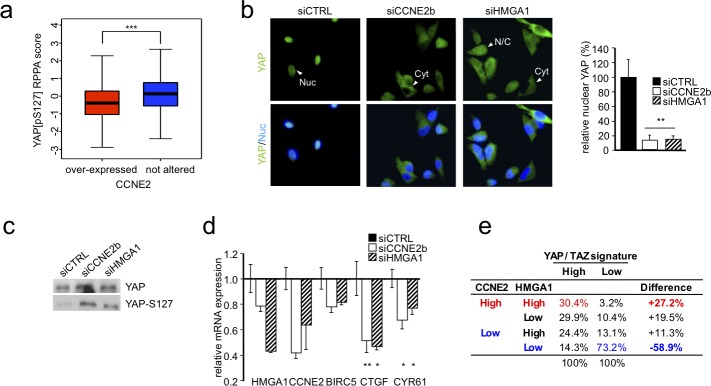
Figure 4. HMGA1 and CCNE2 affect YAP localization and activity.
a. The boxplot shows the YAP protein phosphorylation level in breast cancer samples. The samples were stratified based on the expression level of CCNE2. b. Representative immunofluorescence images of YAP in MDA-MB-231 cells after HMGA1 and CCNE2 silencing are shown. Representative cells are indicated by arrowheads with abbreviations (Nuc, primarily nuclear; Cyt, primarily cytoplasmic; N/C, diffuse in the nucleus and the cytoplasm). Images were taken at X 60 magnification. Right, the ratio of percentage of cells with nuclear YAP treated with siCCNE2b and siHMGA1 compared with siCTRL. The data are presented as the mean±SD (n = 3). c. Western blot analysis of total YAP and YAP phosphorylated at Ser127 (YAP-S127) in protein lysates of MDA-MB-231 cells transfected with siCTRL or siHMGA1 and siCCNE2b (n ≥ 5). d. Real time RT-PCR analysis of YAP target genes (CTGF, BIRC5, CYR61) in MDA-MB-231 cells after HMGA1 and CCNE2 silencing. GAPDH was used for normalization. The data are presented as the mean±SEM (n = 4). e. Contingency table frequencies of breast cancer samples (TCGA dataset) classified as expressing high or low levels of YAP/TAZ profile genes or displaying high or low HMGA1 and CCNE2 expression levels. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed Student's t-test.