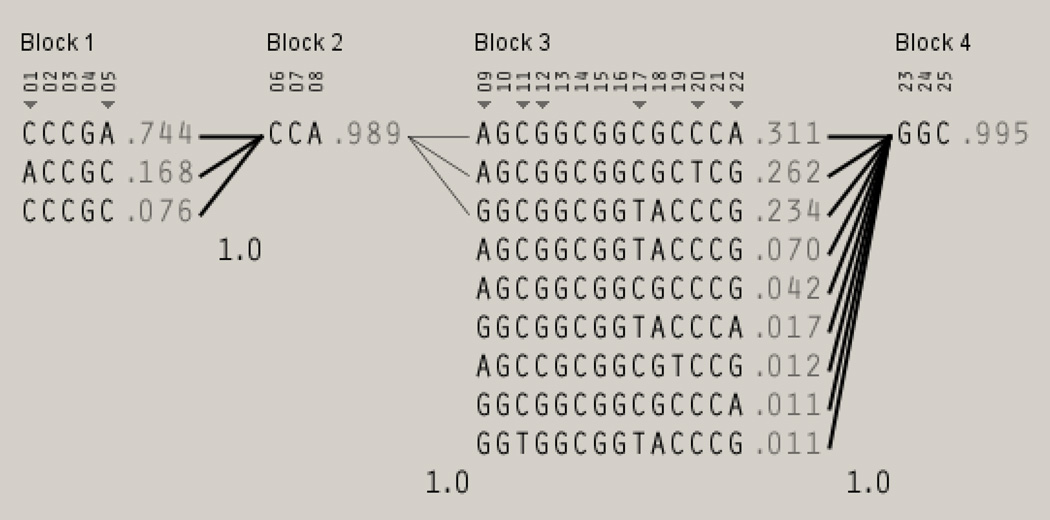
Figure 2. Inferred Haplotypes in Czech Population.
The haplotype display of POR in the Czech cohort (GEVALT v.2. software, GERBIL algorithm) shows each haplotype in a block with its frequency in the respective population and connections from one block to the next (thin and thick lines refer to >10% and >50% chromosomes proceeding from the indicated haplotype on the left to the indicated haplotype on the right). In the crossing areas, a value of multiallelic D' (inversely related to the fraction of chromosomes that have experienced historical recombination) is shown. The numerical labels of the variants correspond to those shown in the Figure 1. Only haplotypes with frequency >1% in the Czech cohort are shown.