Abstract
Epigenetic modifications of DNA and histones alter cellular phenotypes without changing genetic codes. Alterations of epigenetic marks can be induced by exposure to environmental pollutants and may contribute to associated disease risks. Here we test the hypothesis that endothelial cell dysfunction induced by exposure to polychlorinated biphenyls (PCBs) is mediated in part though histone modifications. In this study, human vascular endothelial cells were exposed to physiologically relevant concentrations of several PCBs congeners (e.g., PCBs 77, 118, 126 and 153) followed by quantification of inflammatory gene expression and changes of histone methylation. Only exposure to coplanar PCBs 77 and 126 induced the expression of histone H3K9 trimethyl demethylase jumonji domain-containing protein 2B (JMJD2B) and nuclear factor-kappa B (NF-κB) subunit p65, activated NF-κB signaling as evidenced by nuclear translocation of p65, and up-regulated p65 target inflammatory genes, such as interleukin (IL)-6, C-reactive protein (CRP), intercellular adhesion molecule-1 (ICAM-1), vascular cell adhesion molecule-1 (VCAM-1), and IL-1α/β. The increased accumulation of JMJD2B in the p65 promoter led to a depletion of H3K9me3 repression mark, which accounts for the observed up-regulation of p65 and associated inflammatory genes. JMJD2B gene knockdown confirmed a critical role for this histone demethylase in mediating PCB-induced inflammation of the vascular endothelium. Finally, it was determined, via chemical inhibition, that PCB-induced up-regulation of JMJD2B was estrogen receptor-alpha (ER-α) dependent. These data suggest that coplanar PCBs may exert endothelial cell toxicity through changes in histone modifications.
Keywords: PCBs, epigenetics and vascular inflammation, JMJD2B, H3K9me3, p65, ER-α
Introduction
Epigenetic marks, such as DNA methylation and histone modifications, control gene expression capable of being passed on to daughter cells upon cell division without changing the genetic code [1–3]. Changes in epigenetic marks have been shown to be induced by exposure to various environmental pollutants. Some of these alterations are associated with a broad range of diseases, including cancer, cardiovascular diseases (CVD), or metabolic and reproductive disorders [4–6]. Evidence suggests that persistent organic pollutants such as polychlorinated biphenyls (PCBs), and especially dioxin like (or coplanar) PCBs, can cause epigenetic changes in global DNA methylation, histone modifications in the gene promoters, and expression of corresponding histone modifying enzymes [7–10]. These epigenetic mechanisms may link the environmental exposures to disease outcomes by shaping the genome into active or inactive structures based on endogenous and exogenous environmental changes.
Evidence suggests that the pathology of CVD is associated with exposure to environmental pollutants [11–13]. For example, increased incidence rates of CVD have been demonstrated in populations exposed to high levels of dioxin and PCBs [14–16]. Endothelial dysfunction is believed to be an underlying cause of the initiation of CVD such as atherosclerosis [17]. Exposure to PCBs has also been shown to cause inflammation of the vascular endothelium via expression of several inflammatory markers, cytokines and adhesion molecules [18–20]. Thus, it is of great importance to investigate the underlying mechanisms in the regulation of these inflammatory markers.
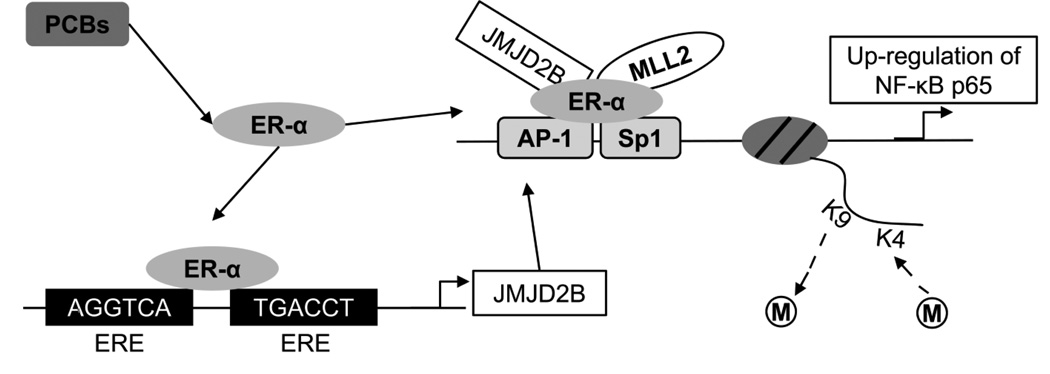
Moreover, PCBs act as endocrine-disrupting chemicals (EDCs), which interfere with the endocrine system by acting as agonists or antagonists to hormone receptors, such as estrogen receptors (ERs), androgen receptors (ARs), or thyroid hormone receptors [21–25]. Previous studies indicate that PCBs have an affinity for ERs and exhibit significant biological activity and disrupt endogenous ER signaling [26–30]. For example, PCB 77 is capable of binding to ER-α and exerting agonistic or antagonistic effects in endothelial cells while PCB 126 exhibits a strong estrogen-like effect by inducing the expression of estrogen receptor α (ER-α) and promoting ER-α-mediated transcription [30–32]. Importantly, ER-α mediates the biological functions of estrogen and regulates transcriptional expression of ER target genes through binding to estrogen responsive elements (EREs) [33, 34]. The jumonji domain-containing protein 2B (JMJD2B), a histone H3K9 trimethyl demethylase, contains two half-ERE sites located in the first intron of its gene, suggesting that JMJD2B itself is transcriptionally targeted by ER-α [35–38]. In addition, JMJD2B is physically associated with ER-α and mixed-lineage leukemia 2 (MLL2), a methyltransferase required for H3K4 trimethylation [38, 39]. It also has been shown that ER-α/JMJD2B/MLL2 complex defines H3K4 and K9 methylation patterns in transcription of target genes [38]. Nuclear factor-kappa B (NF-κB) subunit p65 may be a transcriptional target of ER-α/JMJD2B/MLL2 complex as well, although its promoter does not contain EREs. This transcriptional regulation may be due to ER-α’s ability to interact with regulatory elements of target genes indirectly by associating with activator protein-1 (AP-1) and Sp1 transcription factor complexes and their respective binding sites [33, 34, 40–43]. Importantly, there are consensus binding sites of AP-1 and Sp1 transcription factors in the p65 promoter [44]. Thus, ER-α may function as a mediator in the formation of complexes by histone modifying enzymes and transcription factors. The inflammatory target genes of p65 subunit, such as interleukin (IL-6), C-reactive protein (CRP), intercellular adhesion molecule-1 (ICAM-1), vascular cell adhesion molecule-1 (VCAM-1), IL-1α, and IL-1β [45–55], produce critical inflammatory cytokines that are involved in the initiation and progression of CVD [56]. Taken together, a coherent model has been proposed to implicate PCBs in inflammatory gene regulation through ER-α-mediated epigenetic mechanisms (Fig. 1).
Figure 1. PCB-induced epigenetic regulation of NF-κB subunit p65.
This diagram illustrates the pathways for PCB-induced epigenetic regulation of NF-κB subunit p65. In response to PCBs, ER-α is activated to bind to the EREs in the first intron of JMJD2B and induce its expression. JMJD2B can physically interact with ER-α and MLL2 to form a protein complex and be further recruited to the p65 promoter through AP-1 or Sp1 mediated chromatin binding. This ER-α/JMJD2B/MLL2 complex modifies histone methylation patterns in the promoter region.
The aim of the current study is to investigate the epigenetic mechanisms that link PCBs and vascular inflammation. In particular, we have demonstrated (a) PCB-induced expression of JMJD2B; (b) PCB-activated NF-κB signaling; (c) the role of JMJD2B in PCB-induced p65 transcription; and (d) implication of ER-α in mediating PCB toxicity.
Material and Methods
Cell cultures and treatments
EA.hy926 human endothelial cells (ATCC, Manassas, VA) were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT) as described previously [57]. Cells were grown until confluence and then synchronized by culturing in fresh medium containing 1% serum overnight before treatment.
Stock solution of PCBs (AccuStandard, New Haven, CT) were prepared in DMSO and the same amount of DMSO as in PCB-treated cells were added to control cultures. The concentration of DMSO in the media was kept below 0.05%. Cells were treated with PCBs at concentrations previously found in human plasma (PCBs 77 & 126 at 0.03 nM, PCB 118 at 2 nM, and PCB 153 at 3 nM) [58] for 16 h or up to 24 h for time-course experiments. For ER inhibitor experiments, cells were pre-treated with 0.30 and 0.45 nM of ER inhibitor (Tocris, Minneapolis, MN) for 8 h, followed by treatment with 0.03 nM of PCBs 77 and 126 for 16 h, respectively.
Immunoblotting
Expression of p65 and JMJD2B protein in whole cell extracts was quantified by immunoblot as described previously [19], using mouse monoclonal anti-p65 (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-JMJD2B (Santa Cruz Biotechnology), respectively. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as loading control.
Quantitative real-time PCR (qRT-PCR)
Abundance of mRNA transcripts and amplicons in immunoprecipitated chromatin was evaluated by qRT-PCR (Bio-Rad Laboratories, Hercules, CA) using SYBR Green master mix (Applied Biosystems, Carlsbad, CA) according to the manufacturer’s instructions. The relative amount of each gene was normalized using the housekeeping gene GAPDH. The relative occupancy of epigenetic marks was calculated as described previously [59] and values are reported as fold enrichment. Primer pairs are listed in Table 1.
Table 1.
Oligonucleotide primers used for quantitative real-time PCR
| Gene1 | Sequence (5’-3’) | F/R2 | Template |
|---|---|---|---|
| CRP | GCCCTTCAGTCCTAATGTCCTG | F | cDNA |
| AGCATAGTTAACGAGCTCCCAGA | R | ||
| ER-α | CTTAATTCTGGAGTGTACACAT | F | cDNA |
| CTCCATGCCTTTGTTACTCAT | R | ||
| GAPDH | TCCACTGGCGTCTTCACC | F | cDNA |
| GGCAGAGATGATGACCCTTT | R | ||
| GAPDH | CAAGACCTTGGGCTGGGACTGGCTGA | F | gDNA |
| GATGCGGCTGACTGTCGAACAGGAGGA | R | ||
| ICAM-1 | GCCACCCCAGAGGACAA | F | cDNA |
| CCATTATGACTGCGGCTGCTA | R | ||
| IL-1α | ACAAAAGGCGAAGAAGACTGA | F | cDNA |
| GGAACTTTGGCCATCTTGAC | R | ||
| IL-1β | CTGTCCTGCGTGTTGAAAGA | F | cDNA |
| TTGGGTAATTTTTGGGATCTACA | R | ||
| IL-6 | CAATGAGGAGACTTGCCTGGTGA | F | cDNA |
| TGGCATTTGTGGTTGGGTCAG | R | ||
| JMJD2B | CCAACAGCGAGAAGTACTGTA | F | cDNA |
| CCACGTCGTCATCATACAA | R | ||
| JMJD2B | TCACAGCTGGAATGGTGGT | F | gDNA |
| CACCTCAGGCCCTCAACA | R | ||
| p65 | GGCCATGGACGAACTGTTCC | F | cDNA |
| GAGGGTCCTTGGTGACCAG | R | ||
| p65 | CAGTTTCCCCTCTGGGTGGA | F | gDNA |
| CCCACCTCCCTCCAGAGA | R | ||
| VCAM-1 | GTCTTGGTCAGCCCTTCCT | F | cDNA |
| ACATTCATATACTCCCGCATCCTTC | R |
Genbank entries: CRP = Homo sapiens c-reactive protein (NM_000567.2 for cDNA); ER-α = Homo sapiens estrogen receptor alpha (NM_000125.3 for cDNA); GAPDH = Homo sapiens glyceraldehyde-3-phosphate dehydrogenase (NM_002046.5 for cDNA, NC_000012.11 for gDNA); ICAM-1 = Homo sapiens intercellular adhesion molecule 1 (NM_000201.2 for cDNA); IL-1α = Homo sapiens interleukin-1 alpha (NM_000575.3 for cDNA); IL-1β = Homo sapiens interleukin-1 beta (NM_000576.2 for cDNA); IL-6 = Homo sapiens interleukin-6 (NM_000600.3 for cDNA); JMJD2B (also known as KDM4B) = Homo sapiens lysine (K)-specific demethylase 4B (NM_015015.2 for cDNA, NC_000019.10 for gDNA); p65 (also known as RelA) = Homo sapiens v-rel avian reticuloendotheliosis viral oncogene homolog A (NM_021975.3 for cDNA, NC_000011.9 for gDNA); VCAM-1 = Homo sapiens vascular cell adhesion molecule 1 (NM_001078.3 for cDNA).
Abbreviations: cDNA, complementary DNA (for mRNA quantification); F, forward; gDNA, genomic DNA (for ChIP assays); R, reverse.
Chromatin immunoprecipitation (ChIP)
The enrichment of ER-α, JMJD2B, MLL2, H3K9me3, and H3K4me3 in the promoters of target genes was assessed by ChIP assay as described [60]. Immunoprecipitations were performed with specific antibodies to ER-α (Santa Cruz Biotechnology), JMJD2B (Santa Cruz Biotechnology), MLL2 (Santa Cruz Biotechnology), rabbit IgG (Santa Cruz Biotechnology), H3K9me3 (Abcam, Cambridge, MA), H3K4me3 (Abcam), and H3 (Santa Cruz Biotechnology). Nuclear chromatin extracts without immunoprecipitation were used as input control. Precipitation of chromatin with non-specific IgG was used as a negative control and produced signals much less than those produced by target-specific antibodies. H3 was used as a control that stands for nucleosomal occupancy. The promoter regulating the expression of GAPDH localizes in euchromatin and was used as a control locus. Data are expressed as fold enrichment, representing the ChIP signal as the fold increase in signal relative to the background signal.
Immunocytochemistry
Activation of the NF-κB pathway in endothelial cells was assessed by immunocytochemical localization of p65 subunit. Cells were seeded in 4-well glass chamber slides and treated with coplanar PCBs 77 and 126, or with tumor necrosis factor alpha (TNF-α) (10 ng/ml) as a positive control for 8 h. Cells were washed in phosphate buffered saline (PBS) and fixed in 4% paraformaldehyde for 10 min at room temperature. After washing with PBS, cells were permeabilized with 0.25% Triton X-100 for 10 min. The wash step was repeated, and then cells were blocked with 1% bovine serum albumin in PBS containing 0.1% Tween-20 for 30 min, and subsequently incubated with mouse monoclonal anti-p65 (1:500) overnight at 4°C. After rinsing with PBS, cells were incubated with Alexa Fluor-labeled secondary antibody (1:1000) for 1 h at room temperature in the dark. The 4, 6-diamidino-2-phenylindole (DAPI) was used to stain the nucleus, and the coverslips were mounted onto the slides before viewed under the fluorescence microscope.
JMJD2B gene knockdown
Knockdown of JMJD2B by small interfering RNA (siRNA) (Santa Cruz Biotechnology) was conducted according to the manufacturer’s instructions. Briefly, 1 µl of siRNA duplex was added into 100 µl of transfection medium (solution A) and 6 µl of transfection reagent was diluted into another 100 µl of transfection medium (solution B). Solutions A and B were mixed gently and incubated for 45 min at room temperature. For each transfection, the mixture (solution A &B) was added into 0.8 ml of transfection medium, overlaid onto cells, and incubated at 37° C until confluence before treatment with PCBs.
Statistical analysis
Data were tested to exhibit normal distributions and homogenous variances. Statistical significance was assessed by one-way ANOVA and Fisher’s protected least significant difference posthoc test [61]. JMP 10.0.0 (SAS Institute, Cary, NC) was used to perform all calculations. Data are expressed as means ± SD, and a probability value of p ≤ 0.05 was considered statistically significant.
Results
Coplanar PCBs induce histone demethylase JMJD2B expression
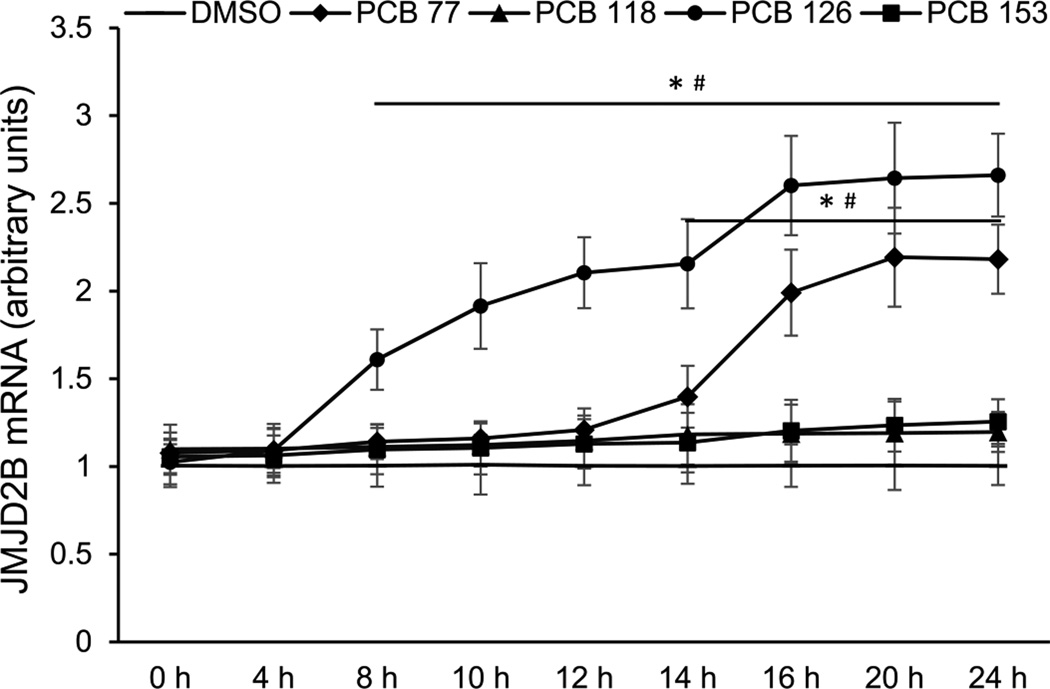
In order to investigate the effects of PCBs on histone modifying enzymes, human endothelial cells were exposed to PCBs 77, 118, 126, and 153 at physiologically relevant levels. JMJD2B is a histone H3K9m3 specific demethylase, which represses transcription by removing methyl groups from K9 on histone H3 tails [36, 38]. The time-course study showed that the expression of JMJD2B was differentially induced by PCBs (Fig. 2). Among the four PCB congeners studied, coplanar PCB 126 most efficiently induced JMJD2B expression, with significant up-regulation observed after 8 h of treatment. Coplanar PCB 77 significantly induced JMJD2B after 14 h of treatment, with the highest induction level observed after 20 h of treatment. Compared to the coplanar PCBs, any apparent up-regulation of JMJD2B expression by PCBs 118 and 153 was non-significant. Thus, in subsequent experiments only coplanar PCBs were used.
Figure 2. Time-course changes of JMJD2B expression in endothelial cells exposed to PCBs.
Cellular JMJD2B mRNA level was measured at indicated time points in endothelial cells following treatments with 0.03 nM of PCBs 77 & 126, 2 nM of PCB 118, or 3 nM of PCB 153. Values are means ± SD (n=3). *Significantly increased compared to the DMSO control. #Significantly increased compare to 0 h.
Coplanar PCBs activate NF-κB signaling via p65 up-regulation and translocation
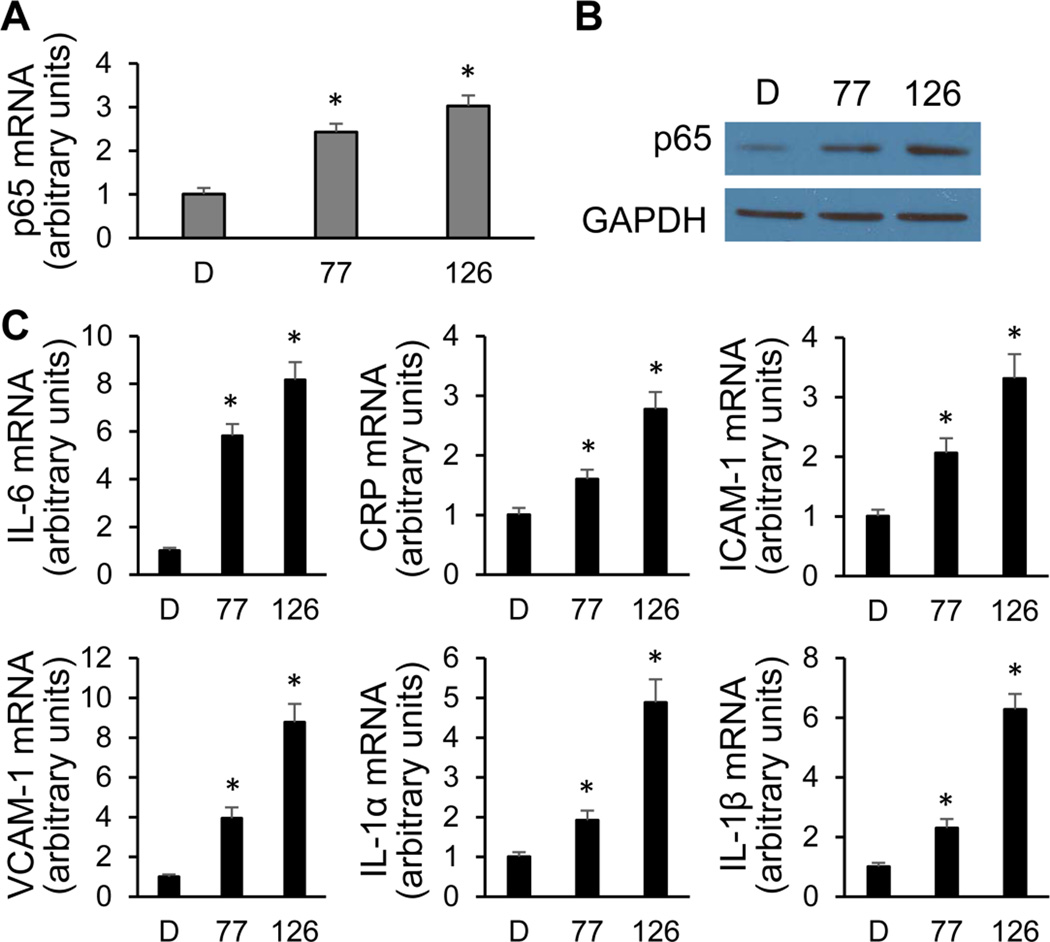
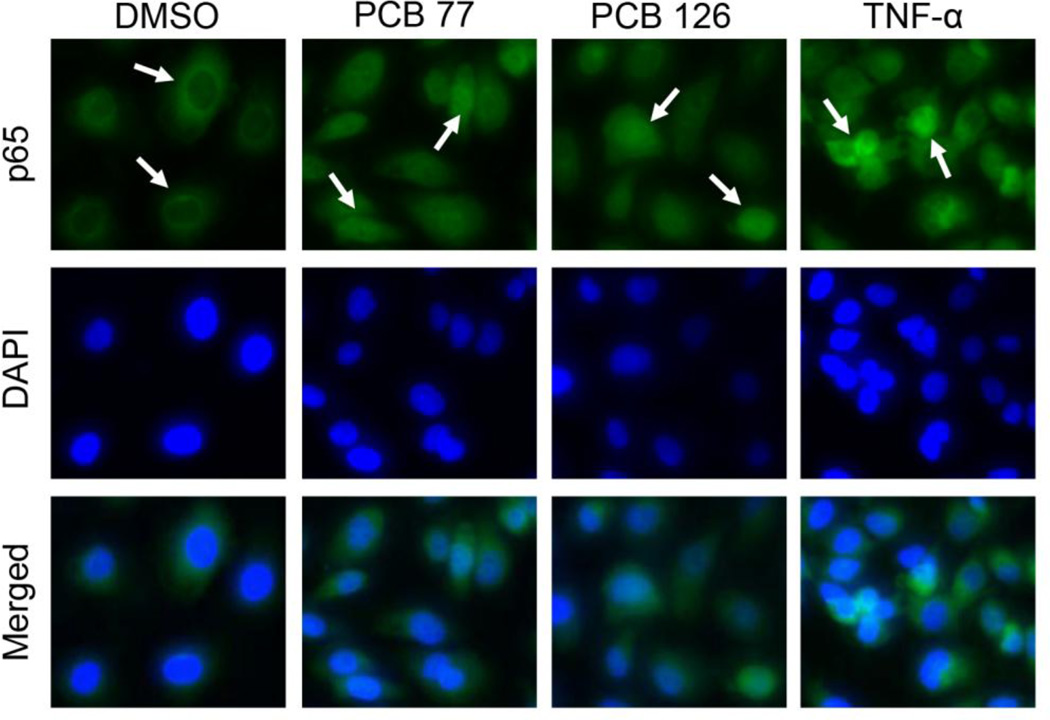
The NF-κB transcription factor is known as a signal integrator that controls two initial steps in the process of vascular inflammation [62]. To test whether PCBs can activate NF-κB signaling, expression of p65, its most prevalent subunit, was measured initially. The expression level of p65 mRNA was increased by approximately 140% and 200% after exposure to PCBs 77 and 126, respectively (Fig. 3A). The increased p65 protein expression as a result of PCB exposure was also detected by immunoblot using whole-cell lysates (Fig. 3B). In addition, expression of p65 target genes IL-6, CRP, ICAM-1, VCAM-1, IL-1α, and IL-1β were also significantly up-regulated after PCB exposure (Fig. 3C). Since the activation of the NF-κB pathway is associated with the intracellular localization of its subunits, p65 nuclear translocation is commonly used as a marker of activation [63]. The intracellular localization of endogenous p65 was analyzed by immunocytochemical staining with anti-p65 antibody, and nuclear DNA was revealed by DAPI staining. The p65 subunit localized to the nucleus after exposure of coplanar PCBs 77 and 126 for 8 h, leading to a substantial increase in the amount of p65 found in the nucleus when compared to the DMSO control (Fig. 4). Cells stimulated with TNF-α were used as a positive control. Taken together, these results suggest that coplanar PCBs activate NF-κB signaling via up-regulating p65 expression and increasing p65 nuclear import in human endothelial cells.
Figure 3. Expression of the NF-κB subunit p65 and its target genes in endothelial cells exposed to PCBs.
(A–B) Coplanar PCBs 77 and 126 induced p65 expression after 16 h of treatment, measured by qRT-PCR and immunoblot. GAPDH is the loading control. (C) Target genes of p65 were up-regulated by coplanar PCBs after 16 h of treatment. Values are means ± SD (n=3). *Significantly increased compared to the DMSO control. D, DMSO; 77, PCB 77; 126, PCB 126.
Figure 4. PCB-induced nuclear translocation of p65 in endothelial cells.
After 8 h exposure with coplanar PCBs, increased nuclear import of p65 was observed in endothelial cells. The intracellular localization of endogenous p65 was analyzed by immunocytochemical staining using anti-p65 (green). The nucleus was visualized by DAPI staining (blue). Nuclear translocation is identified with white arrowheads. Cells were stimulated with TNFα (10 ng/ml) as a positive control.
PCB-induced p65 is transcriptionally activated through histone modifications
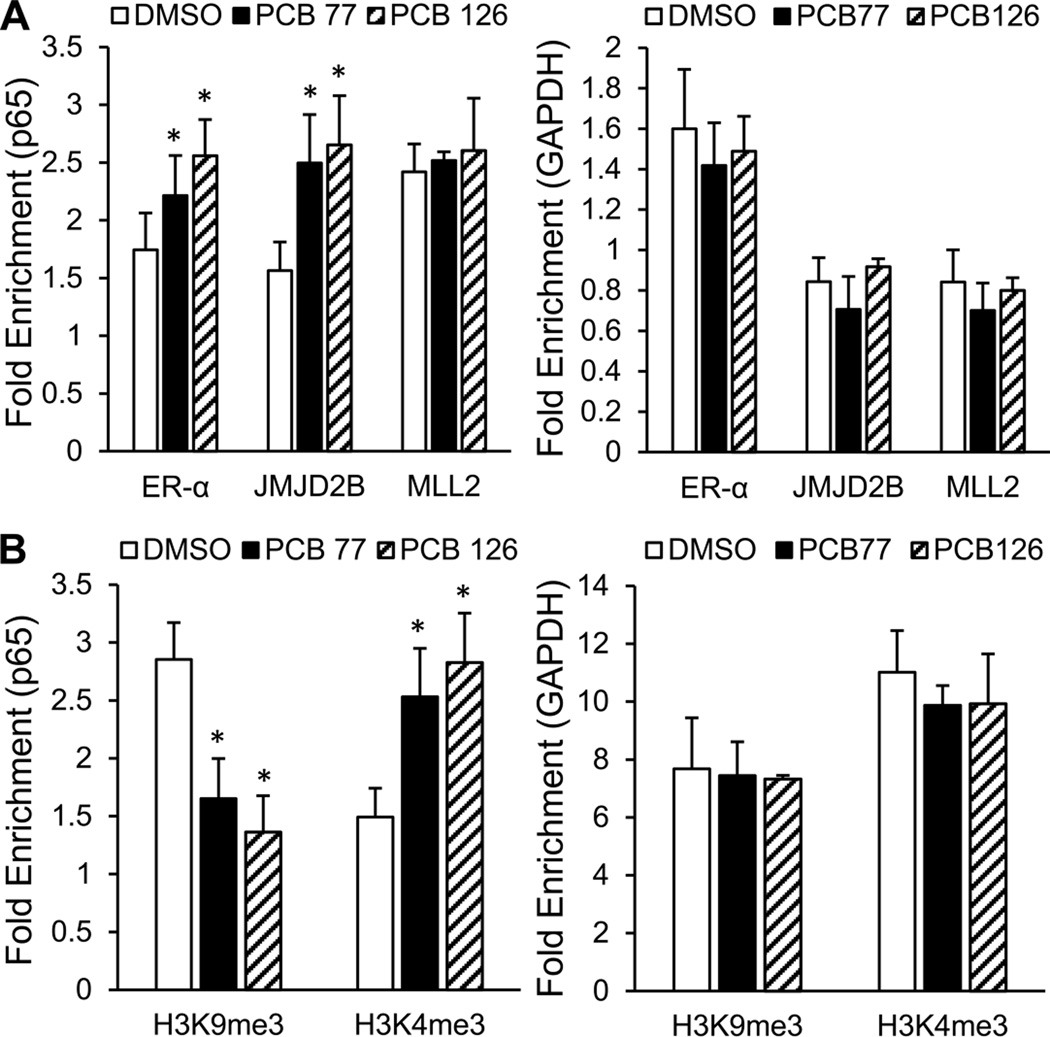
To analyze the role of JMJD2B in p65 transcriptional activation, the accumulation of ER-α/JMJD2B/MLL2 regulatory complex and the enrichment of corresponding histone methylation marks in the p65 promoter were assessed by ChIP assay. Since ER-α may be recruited to the p65 promoter via indirect binding to transcription factors AP-1 or Sp1 [40, 42], the accumulation of ER-α/JMJD2B/MLL2 regulatory complex was assessed within regions that cover AP-1 and Sp1 binding sites. The accumulation of ER-α in the p65 promoter was increased by approximately 27 and 47% after exposed to coplanar PCBs 77 and 126, respectively. The accumulation of JMJD2B was also significantly increased by PCB exposure in a similar pattern. Additionally, the recruitment of MLL2 was slightly enhanced by PCB exposure when compared to the GAPDH control (Fig. 5A). The increased accumulation of JMJD2B and MLL2 in the p65 promoter led to significantly decreased enrichment of H3K9me3 repression marks and significantly increased enrichment of H3K4me3 activation marks, respectively (Fig. 5B). Thus, these data suggest that coplanar PCBs induce p65 expression through JMJD2B-mediated modification of histone methylation.
Figure 5. JMJD2B-mediated modification of histone methylation in the p65 promoter.
(A) The accumulation of ER-α/JMJD2B/MLL2 regulatory complex in the p56 promoter was modulated by PCB exposure. Sheared chromatin from whole-cell lysates was immunoprecipitated with antibodies against the indicated proteins. IgG was used as a negative control. (B) The enrichment of repression mark H3K9me3 and activation mark H3K4me3 in the p65 promoter was changed corresponding to those histone modifying enzymes. H3 is the control for nucleosomal occupancy. The GAPDH promoter was used as control locus. Values are means ± SD (n=3). *Significantly increased compare to the DMSO control.
JMJD2B knockdown abolishes PCB-induced vascular inflammation
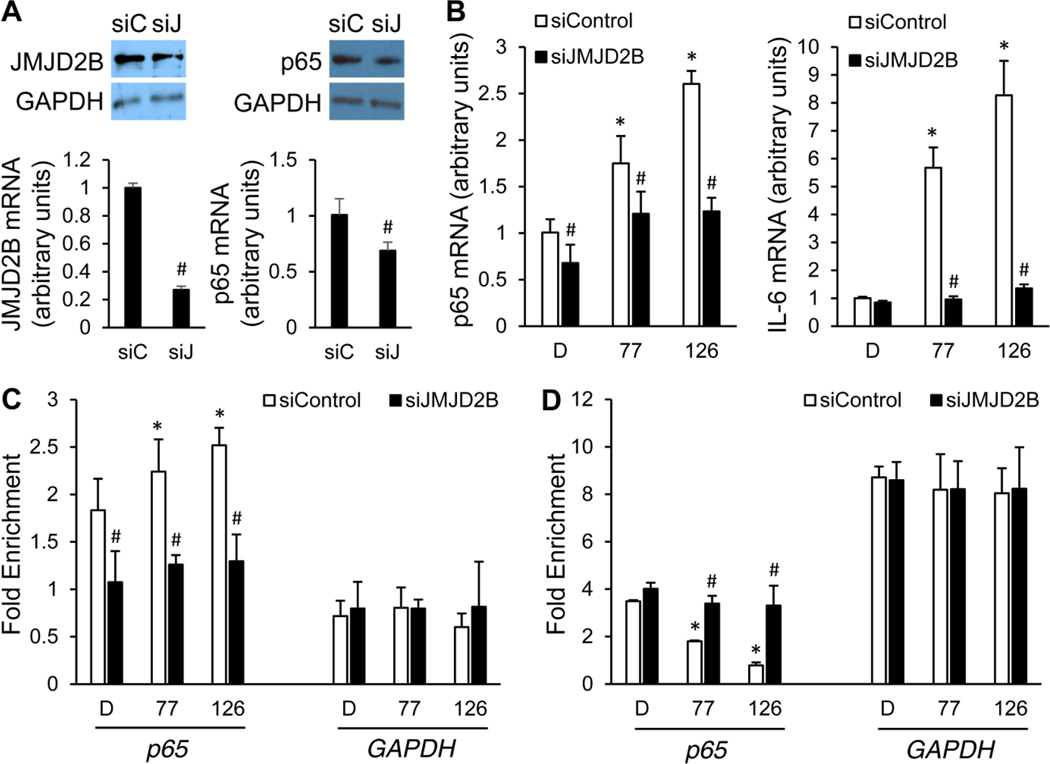
To confirm the important role of JMJD2B in mediating PCB-induced vascular inflammation, knockdown of JMJD2B by siRNA was performed. JMJD2B knockdown was validated at mRNA and protein levels, and it was found that this knockdown significantly reduced p65 expression by 31% (Fig. 6A). Moreover, quantitative RT-PCR revealed that JMJD2B knockdown significantly inhibited the expression of p65 and IL-6 induced by coplanar PCBs 77 and 126 (Fig. 6B). Importantly, since the H3K9me3 mark in the p65 promoter is believed to be modulated by JMJD2B, it was necessary to examine whether depletion of JMJD2B would affect the enrichment of this histone methylation mark in that region. As shown, JMJD2B knockdown significantly inhibited PCB-induced accumulation of JMJD2B in the p65 promoter (Fig. 6C). Consistently, JMJD2B knockdown also abolished PCB-induced removal of H3K9me3 repression mark in the p65 promoter at a significant level (Fig. 6D). Therefore, JMJD2B plays an essential role in mediating PCB-induced inflammation of the vascular endothelium.
Figure 6. Effects of JMJD2B knockdown on PCB-induced vascular inflammation.
(A) JMJD2B knockdown was validated at mRNA and protein levels. Whole-cell lysates from endothelial cells were immunoblotted with antibodies against JMJD2B after 24 h treatment of siRNAs. NF-κB subunit p65 expression was also examined. GAPDH is the loading control. (B) Knockdown of JMJD2B led to significant reduction of PCB-induced p65 and IL-6 expression, measured by qRT-PCR. (C) JMJD2B knockdown reduced the accumulation of JMJD2B in the p65 promoter induced by PCB exposure. (D) JMJD2B knockdown abolished PCB-induced removal of H3K9me3 repression mark in the p65 promoter. *Significantly increased compared to the DMSO control. #Significantly decreased compared to the siRNA control group. siC, siControl; siJ, siJMJD2B; D, DMSO; 77, PCB 77; 126, PCB 126.
ER-α is involved in the regulation of PCB-induced JMJD2B expression
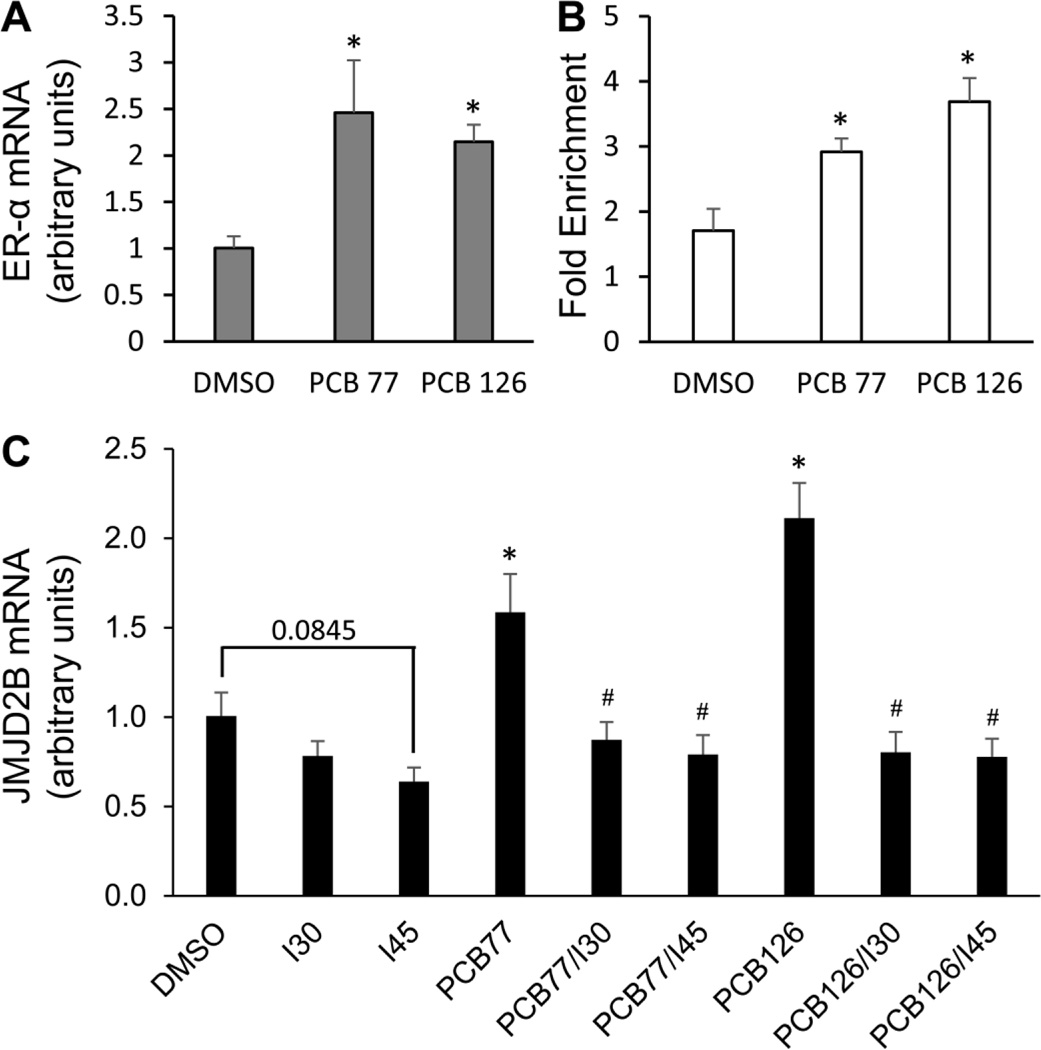
Previous studies suggest that coplanar PCBs have binding affinity to ER-α, induce its expression, and promote ER-α-mediated transcription [30–32]. In endothelial cells, ER-α expression was up-regulated by approximately 146 and 115% after exposed to coplanar PCBs 77 and 126, respectively (Fig. 7A). In addition, there is an ERE site (5’-AGGTCA-NNN-TGACCT-3’) in the first intron of JMJD2B, indicating JMJD2B as a transcriptional target of ER-α [35, 38]. ChIP assay confirmed PCB-dependent recruitment of ER-α to the JMJD2B ERE site in endothelial cells. Coplanar PCBs 77 and 126 significantly increased ER-α binding by approximately 71 and 116 %, respectively compared to the DMSO control (Fig. 7B). In order to test whether ER-α is involved in PCB-induced JMJD2B expression, endothelial cells were pre-treated with ER inhibitor for 8 h and followed by exposure to coplanar PCBs for 16 h. PCB-induced expression of JMJD2B mRNA was significantly attenuated by ER inhibitor at 0.30 and 0.45 nM (Fig. 7C). These data indicate the involvement of ER-α in the regulation of PCB-induced histone demethylase JMJD2B expression.
Figure 7. Involvement of ER-α in the regulation of PCB-induced JMJD2B.
(A) The expression of ER-α was induced by coplanar PCBs 77 and 126 in endothelial cells, quantified by qRT-PCR. (B) The binding of ER-α to the EREs in the first intron of JMJD2B was modulated by PCB exposure. (C) ER inhibitor attenuated PCB-induced expression of JMJD2B mRNA. Cells were pre-treated with ER inhibitor (0.30 and 0.45 nM) for 8 h and followed by treatment of 0.03 nM of coplanar PCBs for 16 h. Values are means ± SD (n=3). *Significantly increased compared to the DMSO control. #Significantly decreased compared to the PCB treatment group.
Discussion
As a chronic inflammatory disease, atherosclerosis is believed to be the primary cause of CVD. Atherosclerotic plaque formation is a dynamic multi-cellular process, which is essentially determined by the regulation of genes in different types of cells [64–66]. The importance of exposure to environmental pollutants in the epigenetic regulation of gene expression during the initiation and progression of atherosclerosis is critical for better understanding of CVD and potential therapeutic interventions based on epigenetic mechanisms. Our study implicates histone methylation modifications in PCB-induced endothelial dysfunctional inflammation, i.e., the early pathology of atherosclerosis, thus providing a potential link between vascular inflammation and epigenetic programming.
Epigenetics provides an attractive explanation of how external factors, e.g., exposure to persistent pollutants like PCBs, can impose aberrant gene expression patterns related to diseases. Epigenetic studies in CVD reveal a significant number of modifications affecting the development and progression of CVD. For example, epigenetics studies of the role of vascular inflammation in patients with CVD show that DNA hypermethylation might be the strongest independent risk factor for CVD mortality [67, 68]. Besides, a recent study investigated the relationship between triple methylation of lysine 27 in histone H3 and atherosclerotic plaque stage, which suggests dynamic alterations in global levels of this methylation mark during atherosclerosis development [69]. Interestingly, epigenetic modifications are reversible and they change rapidly in response to external stimuli such as environmental pollutants. It was reported that persistent pollutants such as PCBs can affect histone modification patterns mediated by histone demethylase Jarid1b [9, 10], indicating that histone modifying enzymes could be potential targets of environmental pollutants.
Our study explored the role of histone demethylase JMJD2B in PCB-induced vascular inflammation. Our data support a model in which coplanar PCBs and JMJD2B participate in the activation of inflammatory genes at the epigenetic level through orchestrating the assembly of a multi-protein histone modifying complex that integrates histone H3K9 demethylation and H3K4 methylation events. In this complex, ER-α is recruited to AP-1 and Sp1 binding sites and further seizes histone modifying enzymes to activate transcription of the NF-κB subunit p65 by changing histone methylation patterns. Recently, the cholesterol metabolite 27-hydroxycholesterol has been identified to be an ER ligand to compete with the action of the atheroprotective estrogen [70, 71]. ER-α also has been reported to be pro-atherogenic because of its role in 27-hydroxycholesterol-promoting atherosclerosis via pro-inflammatory processes [70, 71]. PCBs have been reported to be ER ligands [30, 31], and our study demonstrates that coplanar PCBs can promote endothelial inflammation through the mediation of ER-α, although their competition with estrogen still needs to be confirmed.
In our study, we included the PCB-induced changes in epigenetic marks such as H3K9me3 and H3K4me3 in order to demonstrate a link between epigenetic regulation and the observed PCB-mediated inflammatory response. Histone methylation is usually associated with either activation or repression of transcription, depending on which lysine residue is modified and what histone modifying enzyme is being recruited. For instance, H3K4 methylation marks are always enriched in chromatin adjacent to transcription start sites of active genes, whereas inactive gene promoters are characterized by low levels of H3K4 methylation marks [72]. H3K9 methylation was long considered as a hallmark of heterochromatin, however, it was recently found also to be present in active genes [73–77], suggesting the possible integration or coordination of histone methylation/demethylation in a particular biological process. For example, actively transcribed genes are marked by H3K4me3, and at the same time by H3K9me3, and H3K4 and H3K9 methylation levels are mutually exclusive [78]. Thus, how they coordinate activation of transcription becomes an issue of great importance. Previous studies reported that JMJD2B is physically associated with the MLL2 complex and synergistically shapes the methylation status of H3K9 and H3K4. Notably, H3K9 demethylation is a prerequisite for H3K4 methylation [38]. Our study confirmed that JMJD2B and MLL2 were both recruited to the promoter region of p65 together with ER-α, and the accumulation of this complex was affected by PCB exposure. In particular, we demonstrated that endothelial exposure to coplanar PCBs increased the binding of ER-α/JMJD2B/MLL2 complex to the promoter region of p65, leading to depletion of H3K9me3 marks and thus allowing epigenetic regulation of the NF-κB subunit p65.
In summary, our observations support that JMJD2B plays a significant role in PCB-induced p65 transcription and vascular inflammation through modifying histone methylation marks. The importance of JMJD2B in mediating PCB toxicity was further validated by JMJD2B knockdown studies. With many environmental pollutants contributing to CVD, epigenetic phenomena may hold the key to understanding the onset and progression of this disease. Moreover, histone modifying enzymes such as JMJD2B may also prove to be effective targets for clinical intervention of diseases such as CVD that can be triggered or enhanced by exposure to environmental pollutants.
Highlights.
Coplanar PCBs significantly induced histone demethylase JMJD2B expression.
Coplanar PCBs activated NF-κB through p65 up-regulation and nuclear translocation.
Histone H3K4 and K9 modifications were mediated by ER-α/JMJD2B/MLL2 complex.
ER-α may be involved in the regulation of PCB-induced JMJD2B expression.
Acknowledgements
Research reported in this publication was supported by the National Institute of Environmental Health Sciences at the National Institutes of Health [P42ES007380], National Institute of General Medical Sciences NIH grant 8 P20 GM103527, and the University of Kentucky Agricultural Experiment Station.
Abbreviations
- AP-1
activator protein-1
- ARs
androgen receptors
- ChIP
chromatin immunoprecipitation
- CRP
C-reactive protein
- CVD
cardiovascular disease
- DAPI
4,6-diamidino-2-phenylindole
- EDCs
endocrine-disrupting chemicals
- ERs
estrogen receptors
- ERE
estrogen responsive element
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- H3K4me3
K4-trimethylated histone H3
- H3K9me3
K9-trimethylated histone H3
- ICAM-1
intercellular adhesion molecule-1
- IL
interleukin
- JMJD2B
jumonji domain-containing protein 2B
- K
lysine
- MLL2
mixed-lineage leukemia 2
- NF-κB
nuclear factor-kappa B
- PBS
phosphate buffered saline
- PCBs
polychlorinated biphenyls
- qRT-PCR
quantitative real-time PCR
- siRNA
small interfering RNA
- TNF-α
tumor necrosis factor alpha
- VCAM-1
vascular cell adhesion molecule-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaminsky ZA, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41(2):240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 3.Heijmans BT, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortessis VK, et al. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet. 2012;131(10):1565–1589. doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21(2):243–251. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lind PM, et al. Circulating levels of persistent organic pollutants (POPs) and carotid atherosclerosis in the elderly. Environ Health Perspect. 2012;120(1):38–43. doi: 10.1289/ehp.1103563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desaulniers D, et al. Effects of mixtures of polychlorinated biphenyls, methylmercury, and organochlorine pesticides on hepatic DNA methylation in prepubertal female Sprague-Dawley rats. Int J Toxicol. 2009;28(4):294–307. doi: 10.1177/1091581809337918. [DOI] [PubMed] [Google Scholar]
- 8.Ovesen JL, Schnekenburger M, Puga A. Aryl hydrocarbon receptor ligands of widely different toxic equivalency factors induce similar histone marks in target gene chromatin. Toxicol Sci. 2011;121(1):123–131. doi: 10.1093/toxsci/kfr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casati L, et al. Polychlorinated biphenyls affect histone modification pattern in early development of rats: a role for androgen receptor-dependent modulation? Epigenomics. 2012;4(1):101–112. doi: 10.2217/epi.11.110. [DOI] [PubMed] [Google Scholar]
- 10.Casati L, et al. Androgen receptor activation by polychlorinated biphenyls: epigenetic effects mediated by the histone demethylase Jarid1b. Epigenetics. 2013;8(10):1061–1068. doi: 10.4161/epi.25811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goncharov A, et al. High serum PCBs are associated with elevation of serum lipids and cardiovascular disease in a Native American population. Environ Res. 2008;106(2):226–239. doi: 10.1016/j.envres.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sergeev AV, Carpenter DO. Residential proximity to environmental sources of persistent organic pollutants and first-time hospitalizations for myocardial infarction with comorbid diabetes mellitus: a 12-year population-based study. Int J Occup Med Environ Health. 2010;23(1):5–13. doi: 10.2478/v10001-010-0010-y. [DOI] [PubMed] [Google Scholar]
- 13.Perkins JT, et al. Polychlorinated biphenyls and links to cardiovascular disease. Environ Sci Pollut Res Int. 2015 doi: 10.1007/s11356-015-4479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalton TP, et al. Dioxin exposure is an environmental risk factor for ischemic heart disease. Cardiovasc Toxicol. 2001;1(4):285–298. doi: 10.1385/ct:1:4:285. [DOI] [PubMed] [Google Scholar]
- 15.Gustavsson P, Hogstedt C. A cohort study of Swedish capacitor manufacturing workers exposed to polychlorinated biphenyls (PCBs) Am J Ind Med. 1997;32(3):234–239. doi: 10.1002/(sici)1097-0274(199709)32:3<234::aid-ajim8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 16.Ha MH, Lee DH, Jacobs DR. Association between serum concentrations of persistent organic pollutants and self-reported cardiovascular disease prevalence: results from the National Health and Nutrition Examination Survey, 1999–2002. Environ Health Perspect. 2007;115(8):1204–1209. doi: 10.1289/ehp.10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med. 1999;340(2):115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 18.Eske K, et al. PCB 77 dechlorination products modulate pro-inflammatory events in vascular endothelial cells. Environ Sci Pollut Res Int. 2014;21(10):6354–6364. doi: 10.1007/s11356-013-1591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han SG, et al. Polychlorinated biphenyl-induced VCAM-1 expression is attenuated in aortic endothelial cells isolated from caveolin-1 deficient mice. Toxicol Appl Pharmacol. 2010;246(1–2):74–82. doi: 10.1016/j.taap.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majkova Z, et al. Up-regulation of endothelial monocyte chemoattractant protein-1 by coplanar PCB77 is caveolin-1-dependent. Toxicol Appl Pharmacol. 2009;237(1):1–7. doi: 10.1016/j.taap.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connor K, et al. Hydroxylated polychlorinated biphenyls (PCBs) as estrogens and antiestrogens: structure-activity relationships. Toxicol Appl Pharmacol. 1997;145(1):111–123. doi: 10.1006/taap.1997.8169. [DOI] [PubMed] [Google Scholar]
- 22.Jansen HT, et al. Estrogenic and antiestrogenic actions of PCBs in the female rat: in vitro and in vivo studies. Reprod Toxicol. 1993;7(3):237–248. doi: 10.1016/0890-6238(93)90230-5. [DOI] [PubMed] [Google Scholar]
- 23.Kester MH, et al. Potent inhibition of estrogen sulfotransferase by hydroxylated PCB metabolites: a novel pathway explaining the estrogenic activity of PCBs. Endocrinology. 2000;141(5):1897–1900. doi: 10.1210/endo.141.5.7530. [DOI] [PubMed] [Google Scholar]
- 24.Schrader TJ, Cooke GM. Effects of Aroclors and individual PCB congeners on activation of the human androgen receptor in vitro. Reprod Toxicol. 2003;17(1):15–23. doi: 10.1016/s0890-6238(02)00076-x. [DOI] [PubMed] [Google Scholar]
- 25.Zoeller TR, et al. Thyroid hormone, brain development, and the environment. Environ Health Perspect. 2002;110(Suppl 3):355–361. doi: 10.1289/ehp.02110s3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergeron JM, Crews D, McLachlan JA. PCBs as environmental estrogens: turtle sex determination as a biomarker of environmental contamination. Environ Health Perspect. 1994;102(9):780–781. doi: 10.1289/ehp.94102780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gierthy JF, Arcaro KF, Floyd M. Assessment of PCB estrogenicity in a human breast cancer cell line. Chemosphere. 1997;34(5–7):1495–1505. doi: 10.1016/s0045-6535(97)00446-3. [DOI] [PubMed] [Google Scholar]
- 28.Sumpter JP. Xenoendorine disrupters--environmental impacts. Toxicol Lett. 1998;102–103:337–342. doi: 10.1016/s0378-4274(98)00328-2. [DOI] [PubMed] [Google Scholar]
- 29.Matthews J, et al. Co-planar 3,3',4,4',5-pentachlorinated biphenyl and non-co-planar 2,2',4,6,6'-pentachlorinated biphenyl differentially induce recruitment of oestrogen receptor alpha to aryl hydrocarbon receptor target genes. Biochem J. 2007;406(2):343–353. doi: 10.1042/BJ20070585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelrahim M, et al. 3-Methylcholanthrene and other aryl hydrocarbon receptor agonists directly activate estrogen receptor alpha. Cancer Res. 2006;66(4):2459–2467. doi: 10.1158/0008-5472.CAN-05-3132. [DOI] [PubMed] [Google Scholar]
- 31.Tavolari S, et al. Selected polychlorobiphenyls congeners bind to estrogen receptor alpha in human umbilical vascular endothelial (HUVE) cells modulating angiogenesis. Toxicology. 2006;218(1):67–74. doi: 10.1016/j.tox.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Gjernes MH, Schlenk D, Arukwe A. Estrogen receptor-hijacking by dioxin-like 3,3'4,4',5-pentachlorobiphenyl (PCB126) in salmon hepatocytes involves both receptor activation and receptor protein stability. Aquat Toxicol. 2012;124–125:197–208. doi: 10.1016/j.aquatox.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 33.Robinson-Rechavi M, Escriva Garcia H, Laudet V. The nuclear receptor superfamily. J Cell Sci. 2003;116(Pt 4):585–586. doi: 10.1242/jcs.00247. [DOI] [PubMed] [Google Scholar]
- 34.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29(14):2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 36.Fodor BD, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20(12):1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshioka H, McCarrey JR, Yamazaki Y. Dynamic nuclear organization of constitutive heterochromatin during fetal male germ cell development in mice. Biol Reprod. 2009;80(4):804–812. doi: 10.1095/biolreprod.108.072603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L, et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A. 2011;108(18):7541–7546. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denissov S, et al. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141(3):526–537. doi: 10.1242/dev.102681. [DOI] [PubMed] [Google Scholar]
- 40.Dahlman-Wright K, et al. Interplay between AP-1 and estrogen receptor alpha in regulating gene expression and proliferation networks in breast cancer cells. Carcinogenesis. 2012;33(9):1684–1691. doi: 10.1093/carcin/bgs223. [DOI] [PubMed] [Google Scholar]
- 41.Kushner PJ, et al. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74(5):311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 42.Li C, et al. Requirement of Sp1 and estrogen receptor alpha interaction in 17beta-estradiol-mediated transcriptional activation of the low density lipoprotein receptor gene expression. Endocrinology. 2001;142(4):1546–1553. doi: 10.1210/endo.142.4.8096. [DOI] [PubMed] [Google Scholar]
- 43.Saville B, et al. Ligand-, cell-, and estrogen receptor subtype (alpha/beta)-dependent activation at GC-rich (Sp1) promoter elements. J Biol Chem. 2000;275(8):5379–5387. doi: 10.1074/jbc.275.8.5379. [DOI] [PubMed] [Google Scholar]
- 44.Ueberla K, et al. The NF-kappa B p65 promoter. J Acquir Immune Defic Syndr. 1993;6(3):227–230. [PubMed] [Google Scholar]
- 45.Mori N, Prager D. Transactivation of the interleukin-1alpha promoter by human T-cell leukemia virus type I and type II Tax proteins. Blood. 1996;87(8):3410–3417. [PubMed] [Google Scholar]
- 46.Hiscott J, et al. Characterization of a functional NF-kappa B site in the human interleukin 1 beta promoter: evidence for a positive autoregulatory loop. Mol Cell Biol. 1993;13(10):6231–6240. doi: 10.1128/mcb.13.10.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Son YH, et al. Roles of MAPK and NF-kappaB in interleukin-6 induction by lipopolysaccharide in vascular smooth muscle cells. J Cardiovasc Pharmacol. 2008;51(1):71–77. doi: 10.1097/FJC.0b013e31815bd23d. [DOI] [PubMed] [Google Scholar]
- 48.Libermann TA, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol. 1990;10(5):2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu Y, et al. Costimulation of proliferative responses of resting CD4+ T cells by the interaction of VLA-4 and VLA-5 with fibronectin or VLA-6 with laminin. J Immunol. 1990;145(1):59–67. [PubMed] [Google Scholar]
- 50.van de Stolpe A, et al. 12-O-tetradecanoylphorbol-13-acetate- and tumor necrosis factor alpha-mediated induction of intercellular adhesion molecule-1 is inhibited by dexamethasone. Functional analysis of the human intercellular adhesion molecular-1 promoter. J Biol Chem. 1994;269(8):6185–6192. [PubMed] [Google Scholar]
- 51.Bunting K, et al. Genome-wide analysis of gene expression in T cells to identify targets of the NF-kappa B transcription factor c-Rel. J Immunol. 2007;178(11):7097–7109. doi: 10.4049/jimmunol.178.11.7097. [DOI] [PubMed] [Google Scholar]
- 52.Iademarco MF, et al. Characterization of the promoter for vascular cell adhesion molecule-1 (VCAM-1) J Biol Chem. 1992;267(23):16323–16329. [PubMed] [Google Scholar]
- 53.Zhang D, et al. The effect of interleukin-1 on C-reactive protein expression in Hep3B cells is exerted at the transcriptional level. Biochem J. 1995;310(Pt 1):143–148. doi: 10.1042/bj3100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agrawal A, et al. Overexpressed nuclear factor-kappaB can participate in endogenous C-reactive protein induction, and enhances the effects of C/EBPbeta and signal transducer and activator of transcription-3. Immunology. 2003;108(4):539–547. doi: 10.1046/j.1365-2567.2003.01608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agrawal A, Samols D, Kushner I. Transcription factor c-Rel enhances C-reactive protein expression by facilitating the binding of C/EBPbeta to the promoter. Mol Immunol. 2003;40(6):373–380. doi: 10.1016/s0161-5890(03)00148-2. [DOI] [PubMed] [Google Scholar]
- 56.Incalcaterra E, et al. Pro-inflammatory genetic markers of atherosclerosis. Curr Atheroscler Rep. 2013;15(6):329. doi: 10.1007/s11883-013-0329-5. [DOI] [PubMed] [Google Scholar]
- 57.Lim EJ, et al. The role of caveolin-1 in PCB77-induced eNOS phosphorylation in human-derived endothelial cells. Am J Physiol Heart Circ Physiol. 2007;293(6):H3340–H3347. doi: 10.1152/ajpheart.00921.2007. [DOI] [PubMed] [Google Scholar]
- 58.Schettgen T, et al. Plasma polychlorinated biphenyls (PCB) levels of workers in a transformer recycling company, their family members, and employees of surrounding companies. J Toxicol Environ Health A. 2012;75(8–10):414–422. doi: 10.1080/15287394.2012.674905. [DOI] [PubMed] [Google Scholar]
- 59.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124(1):207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 60.Carey MF, Peterson CL, Smale ST. Chromatin immunoprecipitation (ChIP) Cold Spring Harb Protoc. 2009;2009(9) doi: 10.1101/pdb.prot5279. p. pdb prot5279. [DOI] [PubMed] [Google Scholar]
- 61.Institute S. StatView Reference. 3rd ed. Cary, NC: SAS Publishing; 1998. [Google Scholar]
- 62.Brasier AR. The nuclear factor-kappaB-interleukin-6 signalling pathway mediating vascular inflammation. Cardiovasc Res. 2010;86(2):211–218. doi: 10.1093/cvr/cvq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maguire O, et al. Quantifying nuclear p65 as a parameter for NF-kappaB activation: Correlation between ImageStream cytometry, microscopy, and Western blot. Cytometry A. 2011;79(6):461–469. doi: 10.1002/cyto.a.21068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wierda RJ, et al. Epigenetics in atherosclerosis and inflammation. J Cell Mol Med. 2010;14(6A):1225–1240. doi: 10.1111/j.1582-4934.2010.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hansson GK, Robertson AK, Soderberg-Naucler C. Inflammation and atherosclerosis. Annu Rev Pathol. 2006;1:297–329. doi: 10.1146/annurev.pathol.1.110304.100100. [DOI] [PubMed] [Google Scholar]
- 66.Libby P. Inflammation in atherosclerosis. Nature. 2002;420(6917):868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 67.Castro R, et al. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49(8):1292–1296. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 68.Stenvinkel P, et al. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease? J Intern Med. 2007;261(5):488–499. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- 69.Wierda RJ, et al. Global histone H3 lysine 27 triple methylation levels are reduced in vessels with advanced atherosclerotic plaques. Life Sci. 2015;129:3–9. doi: 10.1016/j.lfs.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 70.Umetani M, et al. The Cholesterol Metabolite 27-Hydroxycholesterol Promotes Atherosclerosis via Proinflammatory Processes Mediated by Estrogen Receptor Alpha. Cell Metab. 2014;20(1):172–182. doi: 10.1016/j.cmet.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Umetani M, Shaul PW. 27-Hydroxycholesterol: the first identified endogenous SERM. Trends Endocrinol Metab. 2011;22(4):130–135. doi: 10.1016/j.tem.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 73.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24(4):800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li F, et al. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008;135(2):272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brinkman AB, et al. Histone modification patterns associated with the human X chromosome. EMBO Rep. 2006;7(6):628–634. doi: 10.1038/sj.embor.7400686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vakoc CR, et al. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26(24):9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vakoc CR, et al. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19(3):381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 78.Binda O. On your histone mark, SET, methylate! Epigenetics. 2013;8(5):457–463. doi: 10.4161/epi.24451. [DOI] [PMC free article] [PubMed] [Google Scholar]