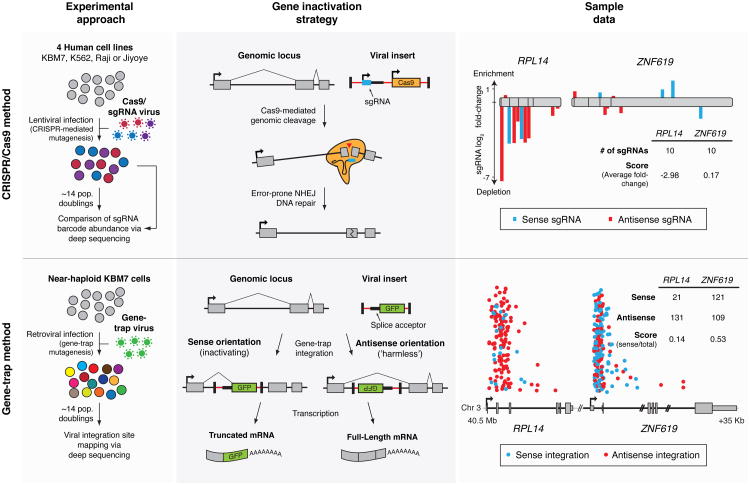
Fig. 1. Two approaches for genetic screening in human cells.
CRISPR/Cas9 (top): Cells are transduced with a genome-wide sgRNA lentiviral library. Gene inactivation via Cas9-mediated genomic cleavage is directed by the 20-bp sequence at the 5′ end of the sgRNA. Cells bearing sgRNAs targeting essential genes are depleted in the final population. Gene-trap (bottom): KBM7 cells are transduced with a gene-trap retrovirus which integrates in an inactivating or “harmless” orientation at random genomic loci. Essential genes contain fewer insertions in the inactivating orientation. Sample data for two neighboring genes RPL14, encoding an essential ribosomal protein, and ZNF619, encoding a dispensable zinc finger protein, are displayed. For CRISPR/Cas9, sgRNAs are plotted according to their target position along each gene with the height of each bar indicating the level of depletion. Boxes indicate individual exons. For gene-trap, the intronic insertion sites in each gene are plotted according to their orientation and genomic position. The height of each point is randomized.