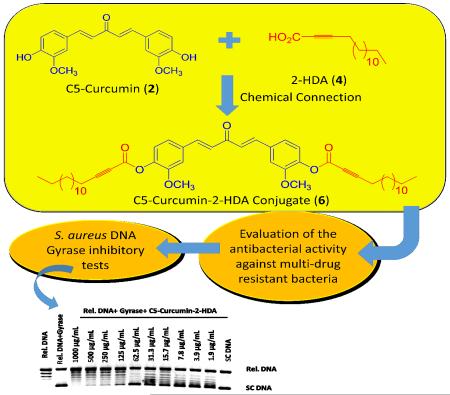
Abstract
The first total synthesis of a C5-Curcumin-2-Hexadecynoic Acid (C5-Curc-2-HDA, 6) conjugate was successfully performed. Through a three-step synthetic route, conjugate 6 was obtained in 13 % overall yield and tested for antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA) strains. Our results revealed that 6 was active against eight MRSA strains at MICs that range between 31.3 and 62.5 μg/mL. It was found that the presence of 2-hexadecynoic acid (2-HDA, 4) in conjugate 6 increased 4-8-fold its antibacterial activity against MRSA strains supporting our hypothesis that the chemical connection of 4 to C5-Curcumin (2) increases the antibacterial activity of 2 against Gram-positive bacteria. Combinational index (CIn) values that range between 1.6 and 2.3 were obtained when eight MRSA strains were treated with an equimolar mixture of 2 and 4. These results demonstrated that an antagonistic effect is taking place. Finally, it was investigated whether conjugate 6 can affect the replication process of S. aureus, since this compound inhibited the supercoiling activity of the S. aureus DNA gyrase at minimum inhibitory concentrations (MIC) of 250 μg/mL (IC50 = 100.2 ± 13.9 μg/mL). Moreover, it was observed that the presence of 4 in conjugate 6 improves the anti-topoisomerase activity of 2 towards S. aureus DNA gyrase, which is in agreement with results obtained from antibacterial susceptibility tests involving MRSA strains.
Keywords: C5-curcuminoids synthesis, antibacterial agents, MRSA, antagonism, DNA gyrase
Graphical Abstract
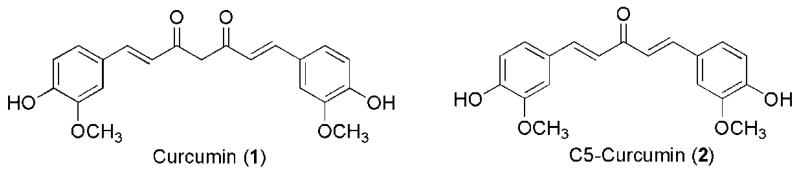
Although our knowledge about infectious diseases produced by multidrug-resistant bacteria has advanced greatly, neither their incidence of spread nor their rate of death has decreased in the last 10 years. The Center for Disease Control and Prevention (CDC) estimated that in the United States, more than 1.2 % out of 2 million people die every year from infections with multidrug-resistant bacteria (MDRB).1 Infectious diseases caused by MDRB add substantial costs to the nation’s health care system since these infections require prolonged and expensive treatments, extended hospitalizations, additional physician visits, and healthcare use, which results in increased rate of disability and death.1 Despite the fact that several compounds are being evaluated as antibacterial agents, it is urgent to develop new antibiotics. Among the compounds that are being evaluated, curcumin (1, Figure 1) has demonstrated to have both anticancer and antibacterial properties.2-6
Figure 1.
Chemical structures of Curcumin (1) and C5-Curcumin (2).
Although preclinical and clinical studies have shown that 1 is not toxic against normal human cells, several pharmacokinetics disadvantages, such as poor bioavailability, fast metabolism and the requirement of repetitive oral doses has been reported, which limits its pharmacological applications.7-9 However, 1 is still an excellent lead compound for the drug design and development of new drugs on the basis of its explicit bioactivities, non-toxicity and easy synthesis.10-12 Several applications have been reported in order to overcome the pharmacokinetics disadvantages mentioned above, which includes the synthesis of bioconjugates of 1 bearing fatty acids13-14, preparation of liposome-encapsulated 115, and synthesis of mono-carbonyl analogues of 1 (C5-curcuminoids).16-18
In 2008, the synthesis of (1E,4E)-1,5-bis(4-hydroxy-3-methoxyphenyl)penta-1,4-dien-3-one (C5-curcumin, 2, Figure 1) was achieved with the goal of evaluating its antibacterial properties.16-17 It was found that 2 was biologically active against Gram-positive and Gram-negative bacteria, including Staphylococcus aureus, Micrococcus luteus, Staphylococcus saprophyticus, Enterococcus sp., Enterobacter cloacae, and Escherichia coli.16 Aimed at improving the antibacterial properties of 1, the synthesis of a curcumin-palmitic acid conjugate (Curc-PA) was performed.14 In this study, Singh et al. demonstrated that the Curc-PA conjugate was five-fold more active against Streptococcus viridans, E. coli, Klebsiella pneumoniae, and Proteus miraleilis than 1 itself. These authors postulated that the enhanced activity of the Curc-PA conjugate is due to the presence of the PA moiety, which decreases the rate of metabolism of curcumin through the hydrolysis of ester bonds by carboesterases present in the cells, provides structural similarity to the cell wall thus allowing a better uptake of 1, enhances its effective concentration, and improves the biological activity of curcumin after enzymatic hydrolysis of the ester group in the Curc-PA conjugate.14 The results mentioned above prompted us to ask whether the chemical conjugation of an antibacterial fatty acid to 2 will improve its antibacterial activity against methicillin-resistant S. aureus (MRSA) strains.
Recently, Sanabria-Ríos and collaborators published the total synthesis of a series of 2-acetylenic fatty acids (2-aFAs) and other synthetic analogs such as 2-tetrahydropyranyl protected alkynols and 2-alkynols aimed at establishing a structure activity relationship (SAR) with these compounds to find the acetylenic fatty acid (aFA) with better antibacterial activity against both Gram-positive and Gram-negative bacteria.19 Among all the compounds tested, 2-hexadecynoic acid (2-HDA, 4) displayed the best overall antibacterial activity against Gram-positive S. aureus (MIC = 15.6 μg/mL), Staphylococcus saprophyticus (MIC = 15.6 μg/mL), and Bacillus cereus (MIC = 31.3 μg/mL), as well as against the Gram-negative Klebsiella pneumoniae (7.8 μg/mL) and Pseudomonas aeruginosa (MIC = 125 μg/mL). In addition, 4 displayed significant antibacterial activity against methicillin-resistant S. aureus (MRSA) ATCC 43300 (MIC = 15.6 μg/mL) and clinical isolates of MRSA (MIC = 3.9 μg/mL). Moreover, we recently determined that 4 is more active than palmitic acid (MIC > 1000 μg/mL) against S. aureus, which demonstrates that the presence of a triple bond at C-2 is important for the antibacterial activity of 4. Taken together, these findings highlight 4 as a novel agent that can be used as a pharmacophore in synthetic applications.
In the present study, the Steglich esterification approach was used to synthesize the C5-curcumin-2-HDA (C5-Curc-2-HDA, 6) conjugate in order to determine its antibacterial activity against Gram-positive and Gram-negative bacteria, including clinical isolates of MRSA (CIMRSA). We hypothesize that by connecting 4 to 2 in 6 its antibacterial activity against MRSA will improve when compared to 2 alone. MRSA strains were selected for this study because the reduction of infections caused by these bacteria in hospital settings represents a priority for the CDC, since these threats are responsible for the largest outbreak of hospital acquired infections.1,20 We are particularly interested in conjugating 4 to 2 because we have demonstrated that this aFA is cytotoxic against several MRSA strains in vitro.19
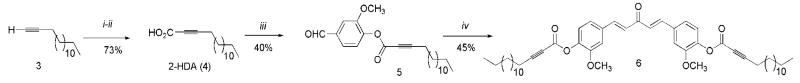
The synthesis of 6 started with the carboxylation of 1-pentadecyne (3) affording 4 in a 73% yield (Scheme 1). Then, 2-HDA was conjugated to vanillin, using Steglich esterification conditions as described in the literature,21-22 affording 5 in a 40% yield. Compound 5 was subsequently reacted with acetone and lithium hydroxide monohydrate to obtain 6 in a 45% yield. The synthetic strategy displayed in Scheme 1 was also used to prepare the C5-Curcumin-Palmitic Acid (C5-Curc-PA) conjugate as described by Sanabria-Ríos et al.22
Scheme 1.
Total synthesis of C5-Curc-2-HDA conjugate (6). i) CO2 (xs.), n-BuLi, THF, −78°C, 24h; ii) 1M HCl, iii) Vanillin, DIC, dry THF-CH2Cl2 (1:1 v/v), DMAP, rt; iv) Acetone, LiOH-H2O, 24 h, rt.
The FT-IR analyses were performed in a Thermo Nicolet IS5 and NMR analyses were performed in a Bruker Avance AV-500 spectrometer. In FT-IR, two bands were observed at 1651 and 1580 cm−1. These bands are generated by one carbonyl group from the ester moiety and the other carbonyl group from the α,β-diunsaturated ketone moiety in 6. In 1H-NMR, two doublets at 7.3 and 6.4 ppm were also observed for 6. These signals have a coupling constant (J) of 20 Hz, which are characteristic of compounds containing a trans alkene moiety.23 Two signals at 188 and 168 ppm were also observed in 13C-NMR, which confirm the presence of the α,β-unsaturated and ester carbonyl groups, respectively.23 The purity of 6 was determined to be > 95% by 13C-NMR. 24
Once 6 was prepared, the conjugate was tested against E. coli (ATCC 25922) and MRSA (ATCC 43300) strains, which were obtained from the American Type Culture Collection (Manassas, VA). In addition, 6 was tested against seven clinical isolates of MRSAs, which were kindly donated from a community hospital in San Juan, Puerto Rico (See Supporting Information). It can be observed from Table 1 that 2 displayed antibacterial activity against eight MRSA strains at MICs that range between 250 and 500 μg/mL, which are in agreement with those results obtained by Liang and collaborators.16 The data presented in Table 1 also demonstrate that the presence of 4 in conjugate 6 increased 4-8-fold the antibacterial activity against MRSA strains supporting our hypothesis that the chemical connection of 4 to 2 increases the antibacterial activity of 2 against Gram-positive bacteria. According to Singh et al., fatty acids are natural components of cell membranes, therefore by conjugating 4 to 2 it is expected that this connection enhances the cellular uptake of 2, increases its lipophilicity, improves its half-life, and reduces its rate of intracellular metabolism by carboesterases, which ensures its low toxicity and high bioavailability.14 It is important to mention, that the antibacterial activity of both palmitic acid (PA) and the C5-Curc-PA conjugate against eight MRSA strains was also tested (Table 1). Results from this test revealed that this conjugate was not active against MRSA strains (MIC > 1,000 μg/mL), which implies that the triple bond at C-2 in 4 has an important role in the antibacterial activity of conjugate 6 against MRSA strains. It can be noted from Table 1 that conjugate 6 is 32-64-fold less active than 4. These results suggest that the addition of a curcuminoid moiety to 4 does not help in increasing its antibacterial activity against Gram-positive bacteria. Taken together these results are in agreement with the findings reported by Sanabria-Ríos et al., where they demonstrated the importance of both the carboxylic acid moiety and the triple bond at C-2 in the antibacterial activity of aFAs against MDRB.19
Table 1.
| Test organism |
MIC/IC50 ± SEM, μg/mL | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 2 | 4 | 6 | PA | C5-Curc- PA |
Methc | Ciprod | |
| MRSA (ATCC 43300) |
250/111.3 ± 10.8 |
7.8/5.1 ± 0.1 |
62.5/39.5 ± 1.9 |
>1000 | >1000 | 1.95/0.98 ± 0.02 |
0.50/0.20 ± 0.09 |
| CIMRSA 1 | 250/141.0 ± 12.4 |
7.8/4.2 ± 0.1 |
62.5/30.3 ± 2.1 |
>1000 | >1000 | 7.8/2.2 ± 0.7 | 15.6/10.4 ± 3.7 |
| CIMRSA 2 | 250/133.4 ± 15.8 |
7.8/5.0 ± 0.5 |
62.5/34.9 ± 1.6 |
>1000 | >1000 | 31.3/11.8 ± 3.9/7.3 |
3.9/2.9 ± 1.2 |
| CIMRSA 3 | 250/123.8 ± 26.2 |
15.6/6.3 ± 0.2 |
62.5/34.2 ± 1.0 |
>1000 | >1000 | 3.9/2.2 ± 0.7 | 7.8/4.4± 0.8 |
| CIMRSA 4 | 500/184.9 ± 8.2 |
15.6/6.5 ± 0.1 |
31.3/24.9 ± 1.5 |
>1000 | >1000 | 31.3/11.8 ± 3.9 |
3.9/2.0 ± 0.2 |
| CIMRSA 5 | 500/202.4 ± 24.5 |
7.8/5.1 ± 0.2 |
31.3/25.4 ± 1.0 |
>1000 | >1000 | 3.9/1.9 ± 0.4 | 15.6/5.3 ± 0.5 |
| CIMRSA 6 | 500/194.7 ± 29.4 |
7.8/5.7 ± 0.8 |
62.5/38.9 ± 1.6 |
>1000 | >1000 | 1.9/1.2 ± 0.1 | 7.8/2.5 ± 0.2 |
| CIMRSA 7 | 500/303.1 ± 33.6 |
7.8/4.7 ± 0.3 |
62.5/34.2 ± 0.4 |
>1000 | >1000 | 3.9/1.3 ± 0.2 | 0.98/0.39 ± 0.04 |
| E. coli (ATCC 25922) |
>1,000 | >1,000 | >1,000 | >1000 | >1000 | >1,000 | <0.008 |
Experiments were performed in triplicate (N =3).
CIMRSA = Clinical isolate of methicillin-resistant S. aureus. CIMRSA stains were kindly donated by a community hospital in San Juan, Puerto Rico.
Meth = Methicillin. Positive control.
Cipro = Ciprofloxacin. Positive control.
The toxicities of 2 and 6 were determined by the CTG Luminescent Cell Viability Assay (See Supporting Information). In addition to Vero cells, the toxicity of 2 and 6 was also determined in peripheral blood mononuclear cells (PBMCs) from healthy volunteers as described by Sanabria-Ríos et al.19,22 The cytotoxicity of 2 and 6 against both Vero cells and PBMCs are displayed in Table 2. Therapeutic Indexes (TIs) of the above-mentioned compounds were calculated as reported by Sanabria-Ríos.19
Table 2.
Cytotoxic activity of 2 and 6 against both normal Vero Cells and PBMC isolated from healthy volunteers.a,b
| Compound | Test MRSA strains | Vero cells cytotoxicity IC50±SEM (μg/mL) |
PBMC cytotoxicity IC50±SEM (μg/mL) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Therapeutic Index (TI)c | ||||||||||
|
| ||||||||||
| ATCC 43300 |
CI 1 | CI 2 | CI 3 | CI 4 | CI 5 | CI 6 | CI 7 | |||
| 2 | 0.4 | 0.3 | 0.4 | 0.4 | 0.3 | 0.2 | 0.2 | 0.2 | 5.7 ± 0.4 | 46.9 ± 0.8 |
| 4 | 13.5 | 16.4 | 13.8 | 10.9 | 10.6 | 13.5 | 12.1 | 14.7 | 1.6 ± 0.4 | 68.9 ± 0.7 |
| 6 | 6.2 | 8.1 | 7.1 | 7.2 | 9.9 | 9.7 | 6.3 | 7.2 | >100.0 | 246.8 ± 12.8 |
Experiments were performed in triplicate (N =3).
CI = Clinical isolate of methicillin-resistant S. aureus.
TI was determined by dividing PBMC cytotoxicity by the inhibitory concentration (IC50) of the drug.
Cytotoxicity results displayed in Table 2 reveal that 2 is toxic against both Vero cells and PBMC at IC50’s of 5.7 and 46.9 μg/mL, respectively. It can be appreciated that 2 is 8.2-fold more toxic against Vero cells than PBMC suggesting that 2 is more toxic towards monkey kidney cells. In terms of TIs, conjugate 6 is 14.7-48.0-fold more therapeutically viable than 2. Results in Table 2 also reveal that 4 is more selective towards MRSA strains, but more cytotoxic against both Vero cells and PBMC, which is in agreement with results obtained by Sanabria-Ríos et al.19 Moreover, these results demonstrate that the chemical connection between 4 and 2 enhance the antibacterial activity of 2 against MRSA strains. The results presented in Table 2 are important because these demonstrate that the conjugation of 2-alkynoic fatty acids can improve the antibacterial properties of C5-curcuminoids and decrease their cytotoxicity against eukaryotic normal cells, which represents a step forward towards the development of a new generation of antibacterial drugs against bacterial infections caused by MDRB.
Results in Table 1 provoked us to ask whether a synergistic effect exists when treating MRSA strains with an equimolar mixture of both 2 and 4. In order to determine synergy, microdilution susceptibility tests were performed using clinical isolates of MRSA (CIMRSA) that were exposed to an equimolar mixture of 2 and 4. Synergism was assessed by constructing dose-response plots and calculating a combinational index (CIn) as described by Chou.25 A CIn < 1 represents a case where synergism between 2 and 4 is present, while CIn values of 1 and > 1 represent additive and antagonistic effects, respectively. Based on the CIn values presented in Table 3, we can appreciate an antagonistic effect when CIMRSA strains were treated with an equimolar mixture of 2 and 4.
Table 3.
Combination index (CIn) values for the equimolar mixture of 2 and 4 in MRSA.a
| Test organism | CIn values |
|---|---|
| MRSA (ATCC 43300) | 2.0 |
| CIMRSA 1 | 2.0 |
| CIMRSA 2 | 2.2 |
| CIMRSA 3 | 1.7 |
| CIMRSA 4 | 1.6 |
| CIMRSA 5 | 1.9 |
| CIMRSA 6 | 2.2 |
| CIMRSA 7 | 2.3 |
CIn vales were determined by using the following equation: , where [2] and [4] are concentrations of 2 and 4 in combination, inhibiting 50% of cell viability, while [2]x and [4]x are the doses of 2 and 4 alone, respectively, inhibiting 50% of cell viability.
Because the equation for calculating CIn assumes that drugs act through an entirely different mechanisms of action25, we theorize that the presence of 2, in an equimolar mixture, may block effectively the active site of important proteins, such as DNA topoisomerases26 and enoyl-ACP reductase InhA27, involved in the antibacterial activity of 4, which could decrease the cytotoxicity of 4 against MRSA strains. Therefore, all of this data strongly support the idea that different mechanisms of action are taking place when CIMRSA are treated with an equimolar mixture of 2 and 4.
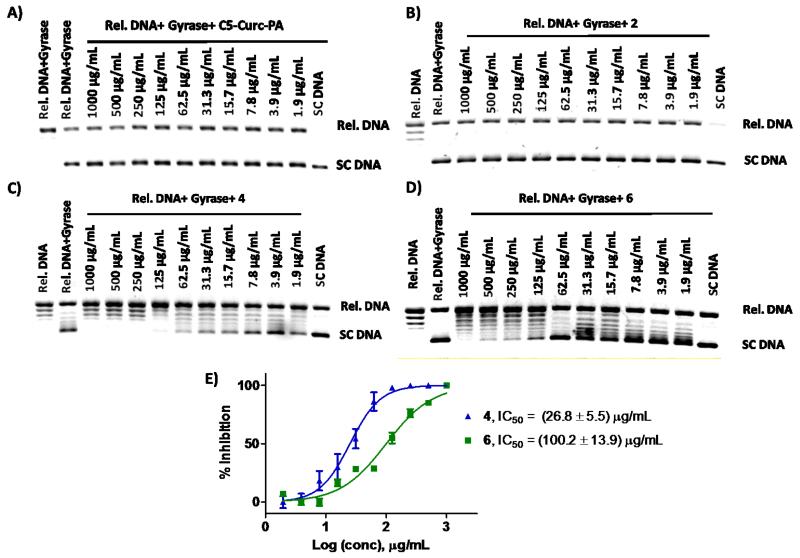
Several studies have demonstrated that curcumin and other curcuminoids display inhibitory activity against DNA topoisomerases I and II.28-30 DNA topoisomerases are enzymes involved in generating the necessary topological and conformational changes in DNA, which are essential for several cellular processes such as replication, recombination, and transcription.31 In addition to their normal cellular functions, DNA topoisomerases are known to be important molecular targets for some anticancer 32-33 and antibacterial drugs.34-36 In order to get more insight regarding the mechanism of action of conjugate 6, we performed several DNA topoisomerase II inhibitory tests by using the S. aureus DNA gyrase kit (TopoGEN, Inc., Port Orange, FL). In addition to 6, the ability of 2 and 4 to inhibit the supercoiling activity of S. aureus DNA gyrase was also tested (See Supporting Information).
The results in Figure 2 show the ability of C5-Curc-PA, 2, 4 and 6 in inhibiting the supercoiling activity of the S. aureus DNA gyrase.
Figure 2.
Representative ethidium bromide stained gels showing the inhibitory activity of C5-Curc-PA (Fig. 2A), 2 (Fig. 2B), 4 (Fig. 2C), and conjugate 6 (Fig. 2D) against the S. aureus DNA gyrase. Dose-response plots showing the inhibitory activity of 4 and 6 against S. aureus DNA gyrase (Fig. 2E). Experiments were performed in triplicate (N =3).
Although these results demonstrate that 4 (Fig. 2C) is 3.7-fold more active against DNA gyrase than 6 (Fig. 2D), the results also support that the chemical connection of 4 to 2 in conjugate 6 enhances the anti-topoisomerase activity of 2 (Fig 2A-D). In addition, it can be appreciated that C5-Curc-PA was not inhibitory against the S. aureus DNA gyrase, which suggests that the presence of a triple bond at C-2 in the fatty acid moiety is important for the anti-topoisomerase activity. Results displayed in Figure 2 are consistent with the findings presented in Table 1, whereby the antibacterial activity of 2 towards MRSAs is improved when connecting compound 4 to 2 in a C5-Curc-2-HDA conjugate. The anti-topoisomerase results in Figure 2, suggest that the presence of 4 in conjugate 6 promotes the interaction of compound 2 with the active site of the S. aureus DNA gyrase resulting in an inhibitory effect of the supercoiling activity of this enzyme, which could affect the DNA replication process in MRSA.
In summary, we successfully performed the total synthesis of conjugate 6 in three synthetic steps and in an overall yield of 13%. Our results clearly demonstrated that the chemical connection of 4 to 2 in conjugate 6 improves both the antibacterial activity of 2 against MRSA strains and its inhibitory effect in inhibiting the supercoiling activity of DNA gyrase, an important enzyme involved in the DNA replication process in bacteria. We are in the process of synthesizing other C5-Curc-aFA conjugates containing different carbon chain lengths, triple bond positions, and degree of unsaturation in order to further explore their antibacterial properties and mechanism of action.
Supplementary Material
Acknowledgements
This project was supported by the National Center for Research Resources and National Institute of General Medical Sciences of the National Institutes of Health the through Grant Number 5 P20 GM 103475-13. J. Rosario acknowledges the support of the PR-INBRE program for an undergraduate fellowship. This project was also supported in part by the PRCTRC Grant through the Grant Numbers U54 RR026139 and 8U54MD 007587-03, and by the National Institute on Minority Health and Health Disparities of the National Institutes of Health through Grant Number 8G12MD007583-28. N. Montano thanks the UPR-Rio Piedras RISE program for a graduate fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1).CDC Antibiotic resitance threat in the United States. Technical Report. 2013:77–78. [Google Scholar]
- 2).Wang Y, Lu Z, Wu H, Lv F. International Journal of Food Microbiology. 2009;136:71–74. doi: 10.1016/j.ijfoodmicro.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 3).Kaur S, Modi NH, Panda D, Roy N. Eur. J. Med. Chem. 2010;45:4209–4214. doi: 10.1016/j.ejmech.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 4).Haukvik T, Bruzell E, Kristensen S, Tonnesen HH. Die Pharmazie. 2011;66:69–74. [PubMed] [Google Scholar]
- 5).Aggarwal BB, Kumar A, Bharti AC. Anticancer Res. 2003;23:363–398. [PubMed] [Google Scholar]
- 6).Aggarwal BB. Cancer Bilogy and Therapy. 2008;7:1436–1440. doi: 10.4161/cbt.7.9.6659. [DOI] [PubMed] [Google Scholar]
- 7).Maheshwari RK, Singh AK, Gaddipati J, Srimal RC. Life Sci. 2006;78:2081–2087. doi: 10.1016/j.lfs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 8).Sharma RA, Steward WP, Gescher AJ. Advances in Experimental Medicine and Biology. 2007;595:453–470. doi: 10.1007/978-0-387-46401-5_20. [DOI] [PubMed] [Google Scholar]
- 9).Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Mol. Pharmacol. 2007;4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 10).Zambre AP, Kulkarni VM, Padhye S, Sandur SK, Aggarwal BB. Bioorg. Med. Chem. 2006;14:7196–7204. doi: 10.1016/j.bmc.2006.06.056. [DOI] [PubMed] [Google Scholar]
- 11).Lin L, Shi Q, Nyarko AK, Bastow KF, Wu CC, Su CY, Shih CC, Lee KH. J. Med. Chem. 2006;49:3963–3972. doi: 10.1021/jm051043z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Handler N, Jaeger W, Puschacher H, Leisser K, Erker T. Chem. Pharm. Bull. 2007;55:64–71. doi: 10.1248/cpb.55.64. [DOI] [PubMed] [Google Scholar]
- 13).Kumar S, Narain U, Tripathi S, Misra K. Bioconjugate Chem. 2001;12:464–469. doi: 10.1021/bc0000482. [DOI] [PubMed] [Google Scholar]
- 14).Singh RK, Rai D, Yadav D, Bhargava A, Balzarini J, De Clercq E. Eur. J. Med. Chem. 2010;45:1078–1086. doi: 10.1016/j.ejmech.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Li L, Braiteh FS, Kurzrock R. Cancer. 2005;104:1322–1331. doi: 10.1002/cncr.21300. [DOI] [PubMed] [Google Scholar]
- 16).Liang G, Yang S, Jiang L, Zhao Y, Shao L, Xiao J, Ye F, Li Y, Li X. Chem. Pharm. Bull. 2008;56:162–167. doi: 10.1248/cpb.56.162. [DOI] [PubMed] [Google Scholar]
- 17).Liang G, Shao L, Wang Y, Zhao C, Chu Y, Xiao J, Zhao Y, Li X, Yang S. Bioorg. Med. Chem. 2009;17:2623–2631. doi: 10.1016/j.bmc.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 18).Yamakoshi H, Ohori H, Kudo C, Sato A, Kanoh N, Ishioka C, Shibata H, Iwabuchi Y. Bioorg. Med. Chem. 2010;18:1083–1092. doi: 10.1016/j.bmc.2009.12.045. [DOI] [PubMed] [Google Scholar]
- 19).Sanabria-Rios DJ, Rivera-Torres Y, Maldonado-Dominguez G, Dominguez I, Rios C, Diaz D, Rodriguez JW, Altieri-Rivera JS, Rios-Olivares E, Cintron G, Montano N, Carballeira NM. Chem. Phys. Lipids. 2014;178:84–91. doi: 10.1016/j.chemphyslip.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Klevens MR, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Graig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 21).Higgins CA, Delbederi Z, McGarel K, Mills T, McGrath O, Feutren-Burton S, Watters W, Armstrong P, Johnston PG, Waugh D, van den Berg H. Bioconjugate Chem. 2009;20:1737–1751. doi: 10.1021/bc900122g. [DOI] [PubMed] [Google Scholar]
- 22).Sanabria-Rios DJ, Rivera-Torres Y, Rosario J, Rios C, Gutierrez R, Carballeira NM, Velez C, Zayas B, Alvarez-Colon F, Ortiz-Soto G, Serrano V, Altieri-Rivera J, Rios-Olivares E, Rodriguez JW. Bioorg. Med. Chem. Lett. 2015;25:2174–2180. doi: 10.1016/j.bmcl.2015.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Lambert JB, Shurvell HF, Lightner DA, Graham Cooks R. Organic Structural Spectroscopy. Prentice Hall; New Jersey: 1998. [Google Scholar]
- 24).Spectral data for 6: 1H-NMR (300 MHz, DMSO-d): δ 7.30 (d, J = 20.10 Hz, 2H), 7.07 (d, J = 8.26 Hz, 2H), 7.00 (m, 2H), 6.90 (m, 2H), 6.40 (d, J = 20.13 Hz, 2H), 3.71 (s, 6H), 2.28 (t, J = 6.89 Hz, 4H), 1.39 (m, 4H), 1.22-0.99 (m, 44H), 0.84 (t, J = 5.97 Hz, 6H); 13C-NMR (75 MHz, DMSO-d): δ 187.57, 167.61, 157.09, 151.04, 130.10, 120.19, 116.82, 107.89, 84.48, 70.90, 68.79, 54.87, 32.10, 31.41, 30.68, 29.62, 29.12, 28.82, 27.53, 23.32, 17.76, 13.96. IR (KBr) νmax: 2960, 2926, 2855, 2237, 1651, 1580, 1539, 1506, 1453, 1399, 1318, 1261, 1218, 1165, 1132, 1125, 1019, 945 cm−1.
- 25).Chou TC. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 26).Carballeira NM, Cartagena M, Sanabria D, Tasdemir D, Prada CF, Reguera RM, Balana-Fouce R. Bioorg. Med. Chem. Lett. 2012;22:6185–6189. doi: 10.1016/j.bmcl.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Morbidoni HR, Vilchèze C, Kremer L, Bittman R, Sacchettini JC, Jacobs WR. Chem. Biol. 2006;13:297–307. doi: 10.1016/j.chembiol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 28).Roth GN, Chandra A, Nair MG. J. Nat. Prod. 1998;61:542–545. doi: 10.1021/np970459f. [DOI] [PubMed] [Google Scholar]
- 29).Ketron AC, Gordon ON, Schneider C, Osheroff N. Biochemistry. 2013;52:221–227. doi: 10.1021/bi3014455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Kumar A, Bora U. Interdisciplinary Sciences. 2014 doi: 10.1007/s12539-012-0048-6. [DOI] [PubMed] [Google Scholar]
- 31).Suzuki K, Shono F, Kai H, Uno T, Uyeda M. J. Enzyme Inhib. 2000;15:357–366. doi: 10.1080/14756360009040693. [DOI] [PubMed] [Google Scholar]
- 32).Giovanella BC, Stehlin JS, Wall ME, Wani MC, Nicholas AW, Liu LF, Silber R, Potmesil M. Science. 1989;246:1046–1048. doi: 10.1126/science.2555920. [DOI] [PubMed] [Google Scholar]
- 33).Sheng C, Miao Z, Zhang W. Current Medicinal Chemistry. 2011;18:4389–4409. doi: 10.2174/092986711797200453. [DOI] [PubMed] [Google Scholar]
- 34).Tse-Dinh YC. Infect. Disord. Drug Targets. 2007;7:3–9. doi: 10.2174/187152607780090748. [DOI] [PubMed] [Google Scholar]
- 35).Bradbury BJ, Pucci MJ. Curr. Opin. Pharm. 2008;8:574–581. doi: 10.1016/j.coph.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 36).Pommier Y, Leo E, Zhang HL, Marchand C. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.