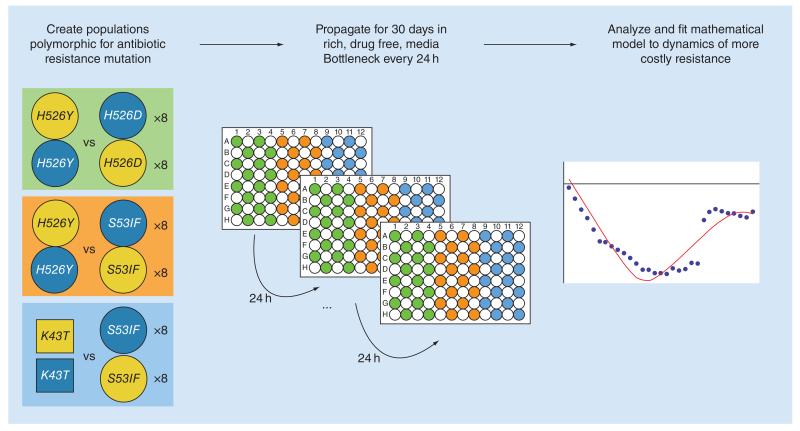
Figure 1. Testing the evolvability of different antibiotic resistances.
First, polymorphic populations are created by mixing, in a 1:1 ratio, clones with two different antibiotic resistance mutations. All clones are isogenic, except for this different resistance mutation and a neutral fluorescent marker. 16 replicate populations are followed for each competition, where half of these replicates have one of the resistances linked with one of the neutral markers, while the other is linked to the other marker. Three different competitive scenarios were studied, between Rifampicin mutations (circles) and between Rifampicin and Streptomycin (squares). Next, populations are passaged in rich media in 96-well plates, organized in a checkered layout and separated by control wells (with media but without bacteria). Every 24 h, all populations are passaged serially into a new 96-well plate with fresh media, during 30 days. The frequencies of the neutral markers (and hence the resistance alleles) are analyzed to unravel possible differences in the evolutionary parameters according to the resistance background.