Abstract
Noroviruses (NoVs) are a leading cause of acute gastroenteritis outbreaks and sporadic cases of diarrhea in industrialized countries. To study the prevalence and genetic diversity of NoVs in Guatemala, stool specimens were collected from hospitalized and ambulatory patients presenting with diarrhea (≥3 loose or liquid stools in a 24-hr period) who were enrolled in a prospective surveillance system in the Departments of Santa Rosa (October 2007 to August 2010) and Quetzaltenango (August 2009 to August 2010), Guatemala. Specimens were tested for rotavirus, enteric bacteria, and parasites by routine methods and for genogroups I and II NoV by real-time reverse transcription-PCR. A total of 2,403 stool specimens were collected from hospitalized (n = 528) and ambulatory patients (n = 1,875). Overall, 341 (14%) samples tested positive for NoVs including 114 (22%) hospitalized and 227 (12%) ambulatory patients. NoVs disease peaked during the winter (November–January) months. Among the 341 NoVs-positive patients, 32 (9%) were also positive for rotavirus, 32 (9%) for bacteria, and 9 (3%) for protozoa. Nucleotide sequences were obtained from 84 samples collected from hospitalized children aged <5 years of age, which could be grouped into nine GII and three GI genotypes with GII.4 (74%) and GI.8 (10%) being the most common. This is the first study on the prevalence of NoVs among hospitalized and ambulatory patients with diarrhea in Guatemala. The findings highlight the need to implement laboratory diagnostics for NoVs to improve appropriate clinical management of diarrheal diseases and guide vaccine development.
Keywords: acute diarrhea, norovirus, surveillance, guatemala
INTRODUCTION
Noroviruses (NoVs) are the leading cause of acute infectious diarrhea and gastroenteritis outbreaks worldwide [Glass et al., 2009]. In developing countries, NoVs are estimated to cause 200,000 deaths each year among children aged <5 years [Patel et al., 2009]. In Latin America data on NoVs are limited, but recent studies suggest they could be important contributors to severe diarrheal disease in children as well as a significant cause of traveler’s diarrhea [Chapin et al., 2005; Bucardo et al., 2008; Gutiérrez-Escolano et al., 2010; Koo et al., 2010; O’Ryan et al., 2010].
NoVs are members of the genus Norovirus within the family Caliciviridae. They can be subdivided genetically into five genogroups (G) of which GI, GII, and GIV are associated with infection in humans [Glass et al., 2009]. In the last decade, GII.4 viruses have emerged as the predominant genotype worldwide responsible for the majority of outbreaks and sporadic cases of norovirus gastroenteritis [Medici et al., 2006; Siebenga et al., 2009; Ferreira et al., 2010; Dai et al., 2011].
There is limited understanding of the epidemiology of NoVs in developing countries, as laboratory diagnosis is not widely available. NoVs are non-cultivable in vitro and, before the 1990s, virus detection depended on demonstration of characteristic viral particles in clinical specimens using electron microscopy. In recent years, the availability of molecular methods such as real-time reverse transcription-polymerase chain reaction (RT-qPCR) and sequence-based typing has allowed a better understanding of the molecular epidemiology of NoVs in developed countries [Vega et al., 2011]. RT-qPCR offers increased sensitivity and specificity with a low risk of cross-contamination, but this capacity is expensive and may not be available in most laboratories in developing countries.
In Guatemala, as in many other countries in Central America, diarrhea is one of the most common causes of morbidity and mortality in children [Bryce et al., 2005], yet little is known about the etiologic role of NoVs. The U.S. Centers for Disease Control and Prevention (CDC), in collaboration with the Guatemalan Ministry of Public Health and Welfare and the University of the Valley of Guatemala (UVG), conducted active facility-based surveillance for diarrheal, respiratory, febrile, and acute infectious neurological diseases in two sites in Guatemala starting in 2007. The present report describes the laboratory-based detection of NoVs among hospitalized and ambulatory patients with diarrhea enrolled in this surveillance system.
MATERIALS AND METHODS
Study Sites
Guatemala is divided into 23 administrative departments. The surveillance data described in this report were collected in the departments of Santa Rosa and Quetzaltenango. The Department of Santa Rosa, located south of Guatemala City, has a population of 308,522 (65% rural and 35% urban) in an area of 3,164 km2. Approximately 97% of the population is mixed European and Amerindian ancestry. Health facilities participating in this study site include Cuilapa Regional Hospital, which serves the entire department, the health center and five health posts that serve the municipality of Nueva Santa Rosa. The Department of Quetzaltenango, located west of Guatemala City, has a population of 780,000 in an area of 2,132 km2, of which 54% are Amerindian indigenous. Health facilities participating in this study site include the Hospital of the Western Region, which serves the entire department, and four health centers (Cantel, Concepción, La Esperanza, and Xecam). A prospective surveillance system with integrated laboratory diagnostics for diarrhea, respiratory disease and unspecified febrile illness was established in Santa Rosa in July 2007 and in Quetzaltenango in February 2009.
Study Patients
An attempt was made to enroll patients of all ages presenting to the surveillance facilities with acute diarrhea, defined as ≥3 liquid stools in a 24-hr period that began during the seven days before presentation. Those who met the case definition and consented were enrolled in the study. For this analysis, we included all patients enrolled from October 2007 to August 2010 in Santa Rosa since the system became fully functional at both hospitalized and ambulatory settings starting in October, and from August 2009 to August 2010 in Quetzaltenango. Of 4,064 eligible diarrhea patients, 3,794 (93%) were enrolled in surveillance, of which 2,567 (68%) provided a stool specimens, of which 2,403 (94%) were tested for norovirus and included in the final analysis. Among the 4,064 eligible patients, the median age was 2 years (range: 1 day to 91 years), with 66% of patients aged <5 years. Likewise, among the 2,403 patients included in the study, the median age was 2 years (range: 1 day to 91 years), with 61% of patients aged <5 years.
Specimen Collection and Processing
Samples of stool obtained by rectal swab were placed in Cary–Blair medium and transported along with bulk whole stool samples at 4°C within 24 hr of collection, to the laboratories at the respective study site hospitals for initial processing. A commercial qualitative EIA was used for the detection of rotavirus (Group A; IDEIA Rotavirus test kits, Dako, Ely, UK). Microbiologic testing included cultures to detect Salmonella spp., Shigella spp., and Campylobacter spp. and microscopic examination to study parasitic pathogens. Stool aliquots were then sent to the UVG laboratories and tested for NoVs by RT-qPCR [Trujillo et al., 2006]. After centrifugation at 6,000 × g for 5 min at 4°C, viral RNA was extracted from 20% clarified stool suspension by using QIAamp Viral RNA Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. A total of 60 μl of purified viral RNA was obtained and stored at −70°C until RT-qPCR analysis.
TaqMan Real-Time RT-PCR
Molecular testing for NoVs was performed as described previously [Vega et al., 2011]. All sets of primers and probes were commercially manufactured (Integrated DNA Technologies, San Diego, CA). For each sample, monoplex RT-qPCR reactions were performed for NoV GI and GII as well as for ribonucleoprotein (RNP) included as an internal control, by using the AgPath-ID One-Step RT-PCR Kit (Applied Biosystems, Foster City, CA) on a 7500 Realtime PCR platform (Applied Biosystems). The amplification of NoV GI was conducted using the primer set Cog 1F/Cog 1R and TaqMan probe Ring 1C [Vega et al., 2011], while NoV GII detection was performed using primer set Cog 2F/Cog 1R and TaqMan probe Ring2 as previously reported [Vega et al., 2011]. The final reaction mix of 25 μl consisted of 0.4 μM of each oligonucleotide primer and 0.2 μM of TaqMan Probe. Cycling conditions included reverse transcription for 10 min at 45°C and denaturation for 10 min at 95°C, followed by 45 cycles of 15 sec at 95°C and 1 min at 60°C. RNP amplification included the sense primer RNP F (5′-AGATTTGGACCTGC-GAGCG-3′), antisense primer RNP R (5′-GAGCGG-CTGTCTCCACAAGT-3′) and TaqMan probe RNP (5′-FAM-TTCTGACCTGAAGGCTCTGCGCG-BHQ-3′). The final reaction mix of 25 μl consisted of 5 μl of the target RNA, 0.30 μM of each primer, 0.10 μM of the probe and water. The RNA was reverse transcribed for 10 min at 45°C followed by 10 min at 95°C and 45 cycles of 15 sec at 95°C, and 1 min at 55°C. Any sample crossing the threshold before cycle threshold (Ct) value of 40 was considered positive.
Sequencing Analysis
Ninety-two NoV positive stool specimens from hospitalized children less than 5 years of age were tested by conventional RT-PCR followed by sequence analysis. All specimens were amplified by conventional RT-PCR for region D [Vinjé et al., 2004] and region D negative samples were amplified for region C [Kojima et al., 2002]. Samples negative for both region C and region D were classified as untypeable. Amplicons were cleaned up by using the QIA Quick Gel Extraction Kit (Qiagen, Valencia, CA) according to the instructions from the manufacturer. Purified DNA was cycle sequenced bi-directionally with Big Dye v1.1 (Applied Biosystems, CA). Cycle sequence clean up was performed with Big Dye Xterminator (Applied Biosystems) and sequenced on a 3130XL Sequencer. Sequences were analyzed on Sequencher 4.7 (Gene Codes, Ann Arbor, MI), BioEdit 7.0 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and phylogenetic analysis done with TreeCon 1.3b (TreeCon, Konstanz, Germany).
Data Analysis
Data were entered during patient interviews using handheld devices. Statistical (Chi-square test, two-sided P-value <0.05 considered significant) and descriptive analyses were performed using SAS v9.2 (SAS Institute, Cary, NC) and Epi Info v3.4.3 (CDC, Atlanta, GA).
Ethics
All patients ≥18 years of age were asked for verbal consent for screening and, if they met the case definition, written, informed consent to participate in the surveillance study. Care providers of children <18 years old were asked for verbal consent to screen their child to determine eligibility, after which written, informed consent was requested from the parents or guardians and written, informed assent from children aged 7 to 17 years old. The study was approved by the Institutional Review Board of the CDC and the UVG (Guatemala City, Guatemala), and by the Guatemalan Ministry of Health and Welfare.
RESULTS
Of 4,064 patients with acute diarrhea that were eligible for participation from Santa Rosa (October 2007 to August 2010) and Quetzaltenango (August 2009 to August 2010), 2,403 (59%) were enrolled and had stool specimens tested for NoVs. Of these, 528 (22%) were hospitalized patients and 1,875 (78%) were from ambulatory clinics. Viral agents were the most frequently identified pathogens among both ambulatory (21%, 385) and hospitalized (49%, 259) patients. NoV was the most common pathogen detected in ambulatory patients (12%), followed by rotavirus (9%), whereas in hospitalized patients, rotavirus was detected most frequently (31%), followed by NoVs (22%; Table I). No etiologic agents were identified in 66% of ambulatory patients and 45% of hospitalized patients.
TABLE I.
Infectious Agent Detected in Stool Specimens From Patients With Diarrhea by Site and Setting (Santa Rosa, Oct 2007 to Aug 2010; Quetzaltenango, Aug 2009 to Aug 2010)
| Santa Rosa
|
Quetzaltenango
|
Both study sites
|
||||
|---|---|---|---|---|---|---|
| Hospital No. (%) n = 458 |
Ambulatory No. (%) n = 1,598 |
Hospital No. (%) n = 70 |
Ambulatory No. (%) n = 1,875 |
Hospital No. (%) n = 528 |
Ambulatory No. (%) n = 1,875 |
|
| Known etiology | ||||||
| Norovirus | 105 (23) | 197 (12) | 9 (13) | 30 (11) | 114 (22) | 227 (12) |
| Rotavirus | 140 (31) | 145 (9) | 22 (31) | 28 (10) | 162 (31) | 173 (9) |
| Bacterial | 41 (9) | 180 (11) | 2 (3) | 10 (4) | 43 (8) | 190 (10) |
| Protozoa | 7 (2) | 103 (6) | 0 (0) | 7 (3) | 7 (1) | 110 (6) |
| Unknown etiology | 198 (43) | 1,031 (65) | 39 (56) | 204 (74) | 237 (45) | 1,235 (66) |
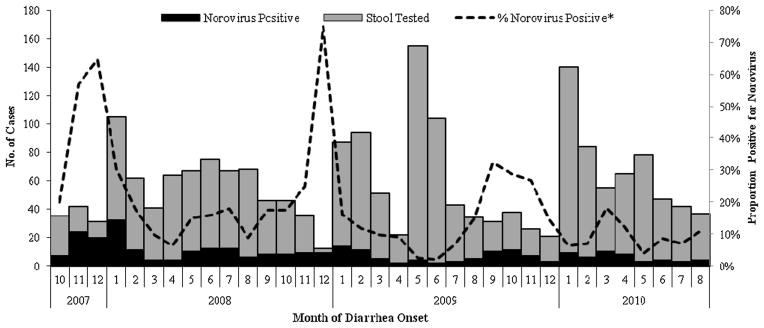
NoV was detected in samples from 341 (14%) patients with diarrhea. The highest number of cases occurred during the colder months of the year (November–January), immediately following the rainy season (Fig. 1). The proportion of stools positive for NoVs during November–January (23%) was significantly higher than that during the other months of the year (11%; P < 0.0001).
Fig. 1.
Monthly distribution of NoVs infection among stool specimens submitted from diarrhea patients, Santa Rosa, Guatemala (Oct 2007 to Aug 2010). *Proportion positive for NoVs (%) was calculated as a function of total number of stool samples tested.
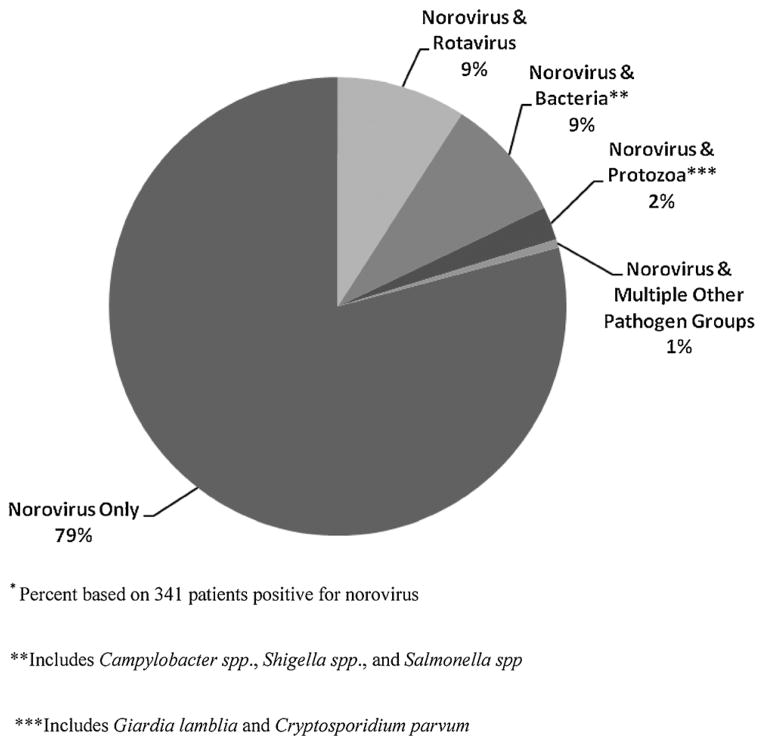
Among the 341 NoV-positive specimens, 281 (84%) tested positive for GII and 53 (18%) for GI, with both genogroups detected in seven (2%) specimens. Mixed infections of norovirus and other enteric pathogens were found in 73 samples, including 32 (9%) specimens positive for rotavirus, 32 (9%) positive for enteric bacteria, and 9 (3%) positive for protozoa (Fig. 2). There was no correlation between coinfection with other diarrhea pathogens and Ct value from a NoV-positive sample.
Fig. 2.
Coinfections in patients positive for norovirus. *Percent based on 341 patients positive for norovirus. **Includes Campylobacter spp., Shigella spp., and Salmonella spp. ***Includes Giardia lamblia and Cryptosporidium parvum.
Ninety-two NoV-positive specimens were selected randomly for further sequencing analysis including 16 GI positive specimens, 72 GII positive specimens, and four specimens that were positive for both GI and GII. Eight (8.7%) samples tested negative for both regions C and D (Table II) and nucleotide sequence analysis of the remaining 84 NoV-positive samples (region D [n = 72] and region C [n = 12]) showed that the most commonly identified genotype was GII.4 Minerva (74%) followed by GI.8 (10%), GII.6 (4%), GII.2 (2%), GII.3 (2%), GII.14 (2%), GI.7 (2%), GI.3 (2%), and GII.13 (1%).
TABLE II.
Norovirus Genotypes Detected Among Hospitalized Children Aged <5 Years in Guatemala; (−) Not Tested
| ORF2 gene
|
|||
|---|---|---|---|
| Region D | Region C | Total | |
| Genogroup I (GI) | n = 12 | n = 12 | |
| GI.8 | 8 | — | 8 |
| GI.3 | 2 | — | 2 |
| GI.7 | 2 | — | 2 |
| Genogroup II (GII) | n = 63 | n = 9 | n = 72 |
| GII.4 Minerva (2006b) | 47 | 1 | 48 |
| GII.4 Riviera (Osaka) | 9 | — | 9 |
| GII.4 Yerseke (2006a) | 2 | 3 | 5 |
| GII.6 | 2 | 1 | 3 |
| GII.3 | 2 | — | 2 |
| GII.2 | 1 | 1 | 2 |
| GII.14 | — | 2 | 2 |
| GII.13 | — | 1 | 1 |
DISCUSSION
This is the first laboratory-based study on the prevalence of NoVs in sporadic cases of diarrhea in Guatemala. The results showed that NoV is one of the leading etiologic agents of acute diarrhea among patients seeking care in Guatemala, with an overall prevalence of 14%, very similar to rotavirus. The greater detection rate of NoVs among hospitalized versus ambulatory patients (22% vs. 12%) underscores further the public health importance of NoV disease.
The NoV prevalence falls within the range reported in other developed and developing countries. Prevalence of severe diarrhea attributable to NoVs reported in other studies ranged from 3% in South Africa and Australia to up to 31% in Peru [Patel et al., 2008]. In children with mild and moderate diarrhea, estimates ranged from 5.5% in Chile to 36% in England [Patel et al., 2008]. Interestingly, Nicaragua has reported a similar prevalence within a pediatric population suggesting that NoVs are a common cause of diarrhea in the Central American region [Bucardo et al., 2008].
NoV was not only the most frequently detected etiological agent in specimens from ambulatory patients, but the virus was, after rotavirus, also the second leading cause of diarrhea requiring hospitalization. Other relevant non-viral enteric pathogens, such as bacteria and protozoa, exhibited lower rates of infections and were found as co-infecting agents in 12% of NoVs-positive samples. In addition, the first laboratory confirmed NoV outbreak in a Guatemala was reported recently [Arvelo et al., 2012] further highlighting the importance of NoVs as a diarrheal pathogen in developing countries.
Sequence analysis of NoV strains in the hospitalized children showed co-circulation of 12 different genotypes. However, GII.4 Minerva was the most common genotype, accounting for approximately three-fourths of the strains. Worldwide, GII.4 is responsible for at least 62% of all NoVs outbreaks [Siebenga et al., 2009] and has also caused multiple NoVs pandemics in the last decade [Bull et al., 2006; Tu et al., 2008; Siebenga et al., 2009]. An experimental vaccine has recently demonstrated proof of concept that NoV illness can be prevented [Atmar et al., 2011], and thus data on the distribution of specific NoVs genotypes in Guatemala and other Latin American countries is critical to guide the development of candidate vaccines effective for the region.
This study has several limitations. First, no parallel enrollment of control patients was performed, so we cannot be certain that detection of NoV was causally associated with illness in all patients. However, we have tested a subset of 60 fecal specimens from healthy children <5 years of age from the Department of Santa Rosa who did not report diarrhea within the 2 weeks preceding interview, and found that only 5% were NoVs GII positive, supporting the etiologic role of NoVs when detected in diarrhea patients (unpublished data). Viral load has been used previously to help discriminate etiologic versus incidental detection of NoV in a fecal sample [Phillips et al., 2009]. Although standard curves using quantified RNA transcripts were not included in the assays and therefore the viral load could not be quantified for each sample, 81% of the NoV positive samples had Ct values lower than 36 suggesting that the majority of the diarrheal illness within the study population could be reasonably ascribed to NoVs infection when detected.
In conclusion, this study demonstrates that NoVs are a major etiologic agent of diarrheal disease in Guatemala. These findings highlight the importance of implementing NoVs diagnostics to assess NoV disease trends and genetic strain diversity over time. This information will not only impact future therapeutic and preventative strategies targeting NoVs infections in Guatemala, but will also be critical in guiding the development and introduction of NoVs vaccines as well as monitoring their subsequent impact.
Acknowledgments
Grant sponsor: CDC; Grant number: UO1 GH000028-02
We acknowledge all the physicians, residents, and nurses at the surveillance hospitals and health facilities. We especially thank Wendy Argueta, Sofía Hernández, Eduviges Molina, and Lucrecia Piloña for their valuable laboratory technical support. We also extend thanks to Gerardo López, Freddy Muñoz, and Karin Ceballos for assisting on database management, and Sharon Roy, Victoria Cuellar, Andrew Thornton, and Jaymin Patel assisted in the collection of samples from healthy children. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC.
Footnotes
None of the authors have any possible conflicts of interest.
References
- Arvelo W, Sosa SM, Juliao P, Lopez MR, Estevez A, Lopez B, Morales-Betoulle ME, Gonzalez M, Gregoricus NA, Hall AJ, Vinjé J, Parashar U, Lindblade KA. Norovirus outbreak of probable waterborne transmission with high attack rate in a Guatemalan resort. J Clin Virol. 2012;55:8–11. doi: 10.1016/j.jcv.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Atmar RL, Bernstein DI, Harro CD, Al-Ibrahim MS, Chen WH, Ferreira J, Estes MK, Graham DY, Opekun AR, Richardson C, Mendelman PM. Norovirus vaccine against experimental human Norwalk virus illness. N Engl J Med. 2011;365:2178–2187. doi: 10.1056/NEJMoa1101245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce J, Boschi-Pinto C, Shibuya K, Black RE. WHO estimates of the causes of death in children. Lancet. 2005;365:1147–1152. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- Bucardo F, Nordgren J, Carlsson B, Paniagua M, Lindgren PE, Espinoza F, Svensson L. Pediatric norovirus diarrhea in Nicaragua. J Clin Microbiol. 2008;46:2573–2580. doi: 10.1128/JCM.00505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RA, Tu ET, McIver CJ, Rawlinson WD, White PA. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J Clin Microbiol. 2006;44:327–333. doi: 10.1128/JCM.44.2.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin AR, Carpenter CM, Dudley WC, Gibson LC, Pratdesaba R, Torres O, Sanchez D, Belkind-Gerson J, Nyquist I, Kärnell A, Gustafsson B, Halpern JL, Bourgeois AL, Schwab KJ. Prevalence of norovirus among visitors from the United States to Mexico and Guatemala who experience traveler’s diarrhea. J Clin Microbiol. 2005;43:1112–1117. doi: 10.1128/JCM.43.3.1112-1117.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai YC, Hu GF, Zhang XF, Song CL, Xiang WL, Wu XB, Wang LY, Jiang X, Nie J. Molecular epidemiology of norovirus gastroenteritis in children in Jiangmen, China, 2005–2007. Arch Virol. 2011;156:1641–1646. doi: 10.1007/s00705-011-1010-3. [DOI] [PubMed] [Google Scholar]
- Ferreira MS, Victoria M, Carvalho-Costa FA, Vieira CB, Xavier MP, Fioretti JM, Andrade J, Volotão EM, Rocha M, Leite JP, Miagostovich MP. Surveillance of norovirus infections in the state of Rio De Janeiro, Brazil 2005–2008. J Med Virol. 2010;82:1442–1448. doi: 10.1002/jmv.21831. [DOI] [PubMed] [Google Scholar]
- Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med. 2009;361:1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Escolano AL, Velázquez FR, Escobar-Herrera J, Lopez Saucedo C, Torres J, Estrada-Garcia T. Human caliciviruses detected in Mexican children admitted to hospital during 1998–2000, with severe acute gastroenteritis not due to other enteropathogens. J Med Virol. 2010;82:632–637. doi: 10.1002/jmv.21743. [DOI] [PubMed] [Google Scholar]
- Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. 2002;100:107–114. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- Koo HL, Ajami NJ, Jiang ZD, Neill FH, Atmar RL, Ericsson CD, Okhuysen PC, Taylor DN, Bourgeois AL, Steffen R, DuPont HL. Noroviruses as a cause of diarrhea in travelers to Guatemala, India, and Mexico. J Clin Microbiol. 2010;48:1673–1676. doi: 10.1128/JCM.02072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici MC, Martinelli M, Abelli LA, Ruggeri FM, Di Bartolo I, Arcangeletti MC, Pinardi F, De Conto F, Izzi G, Bernasconi S, Chezzi C, Dettori G. Molecular epidemiology of norovirus infections in sporadic cases of viral gastroenteritis among children in Northern Italy. J Med Virol. 2006;78:1486–1492. doi: 10.1002/jmv.20723. [DOI] [PubMed] [Google Scholar]
- O’Ryan ML, Peña A, Vergara R, Díaz J, Mamani N, Cortés H, Lucero Y, Vidal R, Osorio G, Santolaya ME, Hermosilla G, Prado VJ. Prospective characterization of norovirus compared with rotavirus acute diarrhea episodes in Chilean children. Pediatr Infect Dis J. 2010;29:855–859. doi: 10.1097/INF.0b013e3181e8b346. [DOI] [PubMed] [Google Scholar]
- Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinjé J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14:1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MM, Hall AJ, Vinjé J, Parashar UD. Noroviruses: A comprehensive review. J Clin Virol. 2009;44:1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Phillips G, Lopman B, Tam CC, Iturriza-Gomara M, Brown D, Gray J. Diagnosing norovirus-associated infectious intestinal disease using viral load. BMC Infect Dis. 2009;9:63. doi: 10.1186/1471-2334-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenga JJ, Vennema H, Zheng DP, Vinjé J, Lee BE, Pang XL, Ho EC, Lim W, Choudekar A, Broor S, Halperin T, Rasool NB, Hewitt J, Greening GE, Jin M, Duan ZJ, Lucero Y, O’Ryan M, Hoehne M, Schreier E, Ratcliff RM, White PA, Iritani N, Reuter G, Koopmans M. Norovirus illness is a global problem: Emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis. 2009;200:802–812. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- Trujillo AA, McCaustland KA, Zheng DP, Hadley LA, Vaughn G, Adams SM, Ando T, Glass RI, Monroe SS. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J Clin Microbiol. 2006;44:1405–1412. doi: 10.1128/JCM.44.4.1405-1412.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu ET, Bull RA, Greening GE, Hewitt J, Lyon MJ, Marshall JA, McIver CJ, Rawlinson WD, White PA. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clin Infect Dis. 2008;46:413–420. doi: 10.1086/525259. [DOI] [PubMed] [Google Scholar]
- Vega E, Barclay L, Gregoricus N, Williams K, Lee D, Vinjé J. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerg Infect Dis. 2011;17:1389–1395. doi: 10.3201/eid1708.101837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinjé J, Hamidjaja RA, Sobsey MD. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J Virol Methods. 2004;116:109–117. doi: 10.1016/j.jviromet.2003.11.001. [DOI] [PubMed] [Google Scholar]