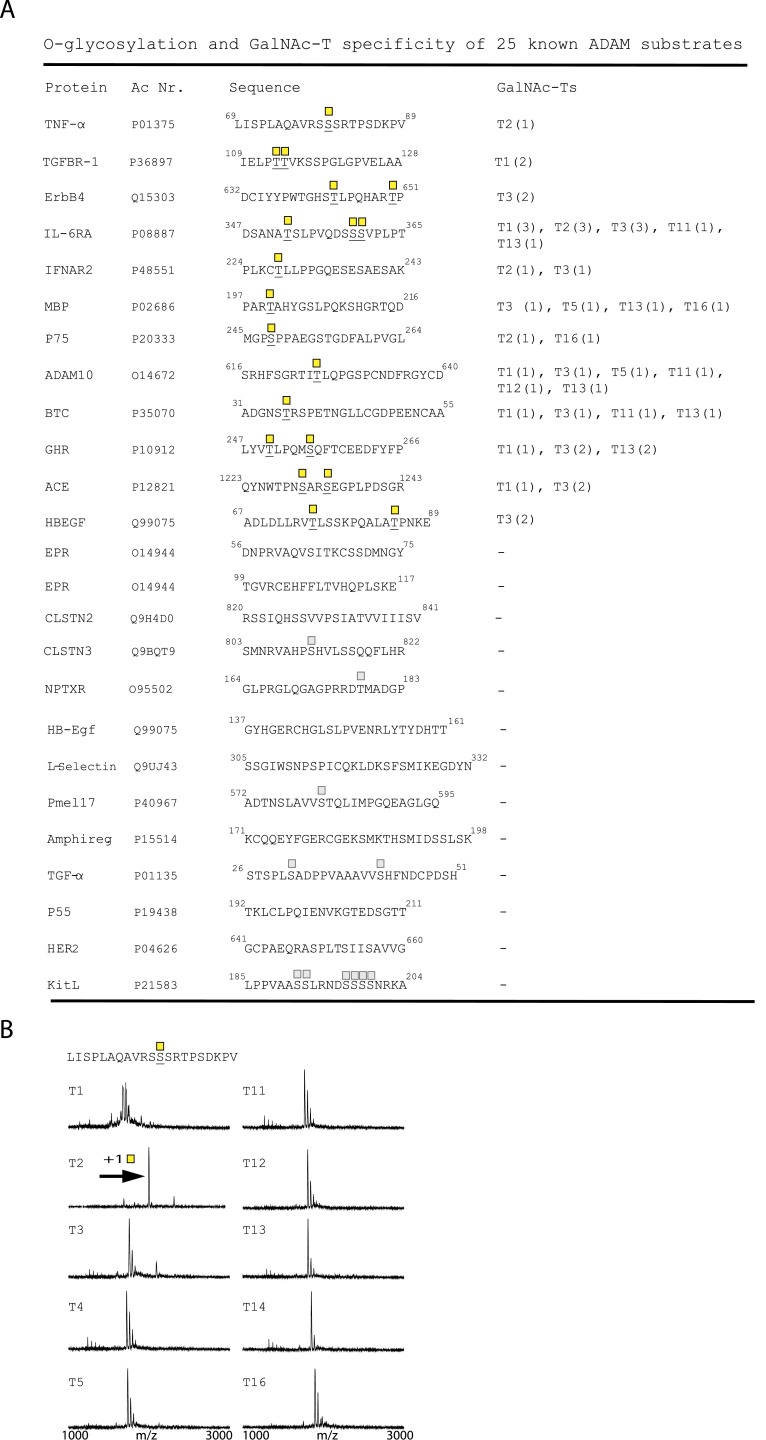
Fig. S1.
In vitro O-glycosylation analysis of known ADAM substrates (A) Peptides of 20–28 residues covering published ADAM cleavage sites. O-glycosylated residues are underlined and indicated by GalNAc (yellow square). In vitro O-glycosylation with GalNAc-T1, -T2, -T3, -T4, -T5, -T11, -T12, -T13, -T14, and -T16. The number of incorporated sites is shown in parenthesis. Predicted (NetOGlyc 4.0) sites in peptides not found to be O-glycosylated are marked by a faded GalNAc. (B) In vitro glycosylation of the juxtamembrane part of TNF-α with 10 different GalNAc-Ts. Horizontal arrows indicate mass shift due to GalNAc glycosylation.