Abstract
To assess the effect of farnesoid X receptor (FXR), a bile acid nuclear receptor, on renal proximal tubular cells, primary cultured mouse kidney proximal tubular cells were treated with GW4064 (a FXR agonist) or DMSO (as controls) overnight. Analysis of gene expression in the proximal tubular cells by whole genome microarrays indicated that FXR activation induced genes involved in fatty acid degradation and oxidation reduction. Among them, genes involved in glutathione metabolism were mostly induced. Here we describe in details the contents and quality controls for the gene expression and related results associated with the data uploaded to Gene Expression Omnibus (accession number GSE70296).
Keywords: Farnesoid X receptor, GW4064, Microarray, Fatty acid metabolism, Oxidation reduction
| Specifications | |
|---|---|
| Organism/cell line/tissue | Mus musculus/kidney/proximal tubular epithelial cells |
| Sex | Male |
| Sequencer or array type | Agilent-026655 Mouse GE 4x44k v2 |
| Data format | Raw data: TXT files, normalized data: TXT |
| Experimental factors | GW4064 vs. DMSO |
| Experimental features | A unique microarray dataset of FXR effect on renal proximal tubular cells |
| Consent | All protocols were approved by the Cantonal Veterinary Office in Zurich. |
| Sample source location | Zurich, Switzerland |
1. Direct link to deposited data
Deposited data can be found here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70296.
2. Experimental design, materials and methods
2.1. Primary culture of mouse kidney proximal tubular cells
Primary proximal tubule cells were isolated from kidneys of C57/BJ mice as described previously [1]. Briefly, kidney cortices from mice were dissected, sliced, minced and digested in 0.25% Trypsin solution (Life Technologies BRL, Grand Island, NY) in a shaking incubator at 37 °C for 1 h. Trypsin was neutralized with growth medium (DMEM and 10% FBS containing 100 U/ml penicillin and 0.1 mg/ml streptomycin). The suspension was pipetted and was passed through a 100-μm cell strainer (Becton Dickinson Labware, Franklin Lakes, NJ). The samples were centrifuged (600 rpm for 5 min) to pellet the tubules, washed with 10 ml of medium, centrifuged, and washed twice more. The final pellet, consisting mostly of renal tubules, was resuspended in culture medium (REBM bullet kit, Clonetics), plated onto culture dishes (Nalge Nunc International, Naperville, IL) and incubated at 37 °C in a carbon dioxide incubator with medium changes every 2 days until confluent. Experiments were carried out in serum-free DMEM. To activate FXR, cells were starved for 12 h in 0.2% FBS DMEM, then incubated by the addition of 1 μM GW4064 (Sigma-Aldrich, St. Louis, MO USA), or DMSO (as controls) overnight.
2.2. Microarray and gene expression analysis
RNA was extracted from primary cultured mouse proximal tubule cells using RNeasy Microarray Tissue mini kit (73304, Qiagen, Germany), followed by on column DNase digestion to remove any contaminating genomic DNA. RNA samples from 4 dishes per group were subjected to microarray analysis. Briefly, 100 ng of total RNA was reverse-transcribed into double-stranded cDNA, which was linearly amplified and labeled with Cy3 dye. Following quantification using a Nanodrop spectrophotometer (Witec, Luzern, Switzerland) and quality assessment with Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), 1.6 μg of the obtained Cy3-labeled cRNA was hybridized to Mouse GE 4x44K v2 Microarrays (Agilent Technologies, Santa Clara, CA) according to the manufacturer's protocol. Arrays were scanned with an Agilent G2565CA Microarray Scanner System (Agilent, Santa Clara, CA). Raw intensity data were obtained using Agilent's Feature Extraction Software version 10.7 for array image analysis and the calculation of spot intensity measurements. All microarray data were submitted to the Gene Expression Omnibus [2] (accession number GSE70296).
2.3. Normalization
Data analysis was carried out with R/Bioconductor [3]. The processed intensities and normalized across samples were loaded by using quantile normalization. All microarray data was submitted to the Gene Expression Omnibus (accession number GSE70296). Differential expression was computed using the limma package [4], [5]. More details on analysis methods can be found at http://fgcz-bfabric.uzh.ch/wiki/tiki-index.php?page=app.two_groups. Gene oncology analysis, network analysis and KEGG pathway analysis of the microarray data were completed using the DAVID Bioinformatics Resources (National Institute of Allergy and Infectious Diseases, NIH, USA).
2.4. Basic analysis
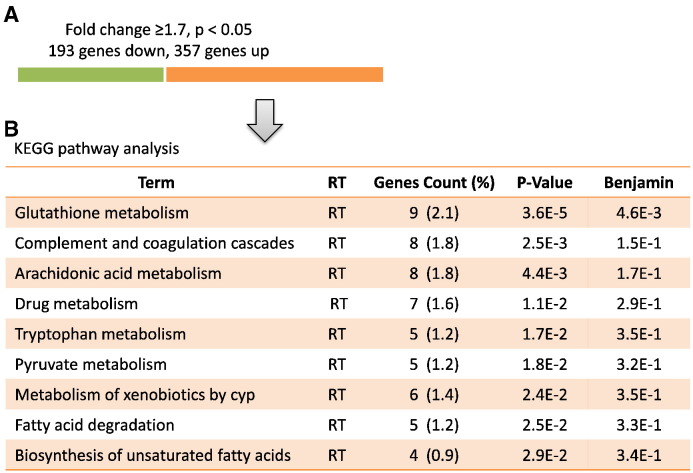
First, gene expression was compared between GW4064-treated cells versus DMSO-treated cells. By using a ≥ 1.7-fold change as a cut-off, 550 genes were significantly expressed by FXR activation. Among them, 193 genes were down-regulated and 357 genes were up-regulated (Fig. 1A). Interestingly, by KEGG pathway analysis, several pathways involved in fatty acid metabolism and oxidation reduction were induced by FXR activation. Pathways belonging to oxidation reduction include glutathione metabolism pathway, drug metabolism pathway and xenobiotic metabolism pathway (Fig. 1B). Furthermore, by comparison between public available ChIP-seq data (GSE57305, altered mRNA expression profile of GW4064 treated mouse livers compared to vehicle control, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57305) with our microarray data (GSE70296, effect of GW4064 on primary cultured mouse kidney proximal tubule cells), we identified that activated FXR could regulate similar pathways in mouse liver in vivo as the pathways in mouse proximal tubular cells (such as glutathione metabolism pathway, drug metabolism pathway, fatty acid metabolism pathway and pyruvate metabolic process pathway), indicating a universal role of FXR in both mouse kidney and liver (Table 1).
Fig. 1.
Comparison of distinct gene expression patterns between the different experimental conditions. (A) Summary of differentially expressed genes between GW4064 treatment overnight and vehicle control (DMSO). Genes that are differentially increased or decreased after GW4064 treatment are indicated by orange and green bars, respectively. Cut-off 1.7-fold, p < 0.05. Of all 550 genes significantly expressed after GW4064 treatment, 357 genes are increased and 193 genes are reduced. (B) KEGG pathway analysis of genes with a change in expression of ≥ 1.7 fold increase (orange bars).
Table 1.
| Top pathways induced by FXR in proximal tubular cells (PTCs-GW from GSE70296) |
Top pathways induced by FXR in mouse liver (mLiver-GW from GSE70296) |
||
|---|---|---|---|
| Name | p value | Name | p value |
| Glutathione metabolism | 3.6E − 5 | Pyruvate metabolism | 5.4E − 5 |
| Complement and coagulation | 2.5E − 3 | Drug metabolism | 1.6E − 4 |
| Arachidonic acid metabolism | 4.4E − 3 | PPAR signaling pathway | 6.2E − 4 |
| Drug metabolism | 1.1E − 2 | Fatty acid metabolic process | 8.2E − 4 |
| Tryptophan metabolism | 1.7E − 2 | Glutathione metabolism | 2.5E − 3 |
| Pyruvate metabolism | 1.8E − 2 | Complement and coagulation | 3.3E − 3 |
| Metabolism of xenobiotics | 2.4E − 2 | Metabolism of xenobiotics | 3.1E − 2 |
| Fatty acid degradation | 2.5E − 2 | Retinol metabolism | 2.2E − 2 |
| Biosynthesis of unsaturated fatty acids | 2.9E − 2 | Biosynthesis of unsaturated fatty acids | 1.7E − 3 |
Similar top pathways induced by activated FXR in both datasets are highlighted in bold.
3. Discussion
We described here a unique mRNA profile of GW4064 treated mouse kidney proximal tubular cells. This dataset is composed of genome-wide expression profiling data measured by Agilent platform. We showed that FXR activation by GW4064 induced genes involved in fatty acid metabolism and oxidation reduction. Among them, glutathione metabolism pathway was mostly induced by FXR activation.
References
- 1.Gai Z., Zhou G., Gui T., Itoh S., Oikawa K., Uetani K., Muragaki Y. Trps1 haploinsufficiency promotes renal fibrosis by increasing Arkadia expression. J. Am. Soc. Nephrol. 2010;21:1468–1476. doi: 10.1681/ASN.2009121201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., Kim I.F., Soboleva A., Tomashevsky M., Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quackenbush J. Microarray data normalization and transformation. Nat. Genet. 2002;32:496–501. doi: 10.1038/ng1032. (Suppl) [DOI] [PubMed] [Google Scholar]
- 5.Smyth G.K. Springer; New York: 2005. limma: Linear Models for Microarray Data. [Google Scholar]