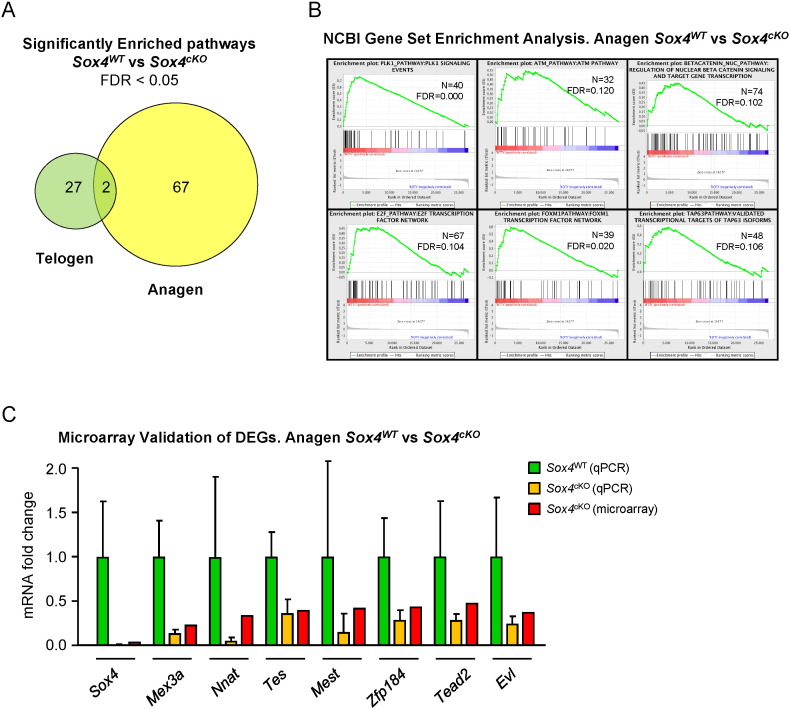
Fig. 1.
A) Venn's diagram showing the overlap between the significantly enriched pathways in Sox4WT vs Sox4cKO mouse skin in Telogen (green) and Anagen (yellow). Only pathways showing FDR < 0.05 were included for this comparison. Note that most deregulated pathways belong to the Anagen group, reinforcing a role for Sox4 in this condition, and that the overlap is minimal between both categories.
B) Enrichment plots for selected pathways, obtained from the NCI repository at FDR < 0.150, where N indicates the number of genes included in each list, and FDR means false discovery rate (q value). The red to blue bar represents the ranked list of genes (red = Sox4WT, blue = Sox4cKO). Genes showing differential expression between genotypes are located at the edges of the bar, and similarly-expressed genes are in the centre. The Y axis represents the enrichment score (ES). Plk1, ATM, nuclear β-catenin, E2F, FoxM1 and TAP63 pathways are shown from left to right and top to bottom.
C) Comparison of the values obtained by qPCR and microarray in Sox4WT vs Sox4cKO mouse skin, for the selected DEGs. Fold change relative to Sox4WT (set to 1) is shown. N = 6 mice per genotype.