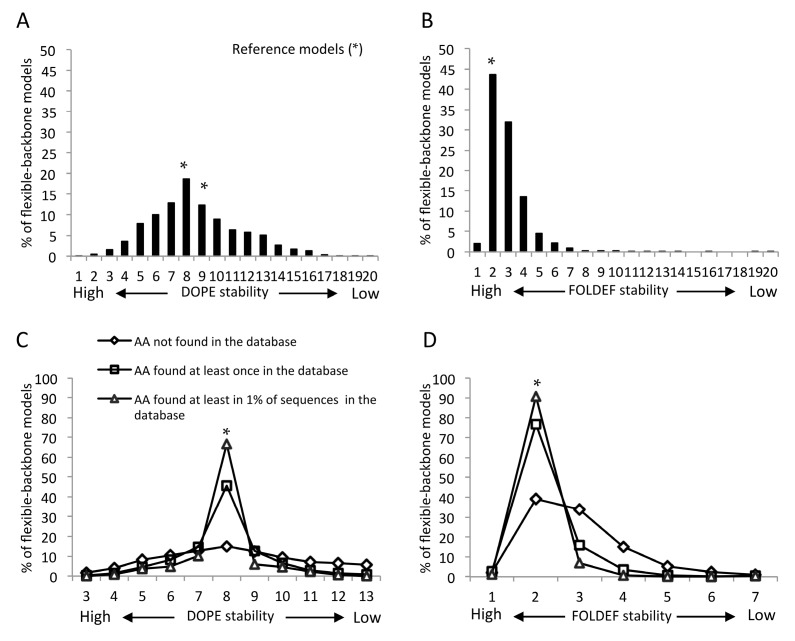
Figure 1.
Distribution of capsid protein mutant stabilities based on flexible-backbone models of the mature capsid (CA) hexamer. The stability bin reflects the structural stability from higher (left) to Revise the asterisks into Palatino linotype. lower (right) levels. * indicates the bin in which reference structures were found. All mutations predicted by Discrete optimized protein energy (DOPE) (A,C) and FoldX energy function (FOLDEF) (B,D) were classified into three groups based on their frequency in the HIV sequence database. Only results from five higher, five lower and the reference model bins are shown, as together they accounted for more than 98% of all models.