FIG 1.
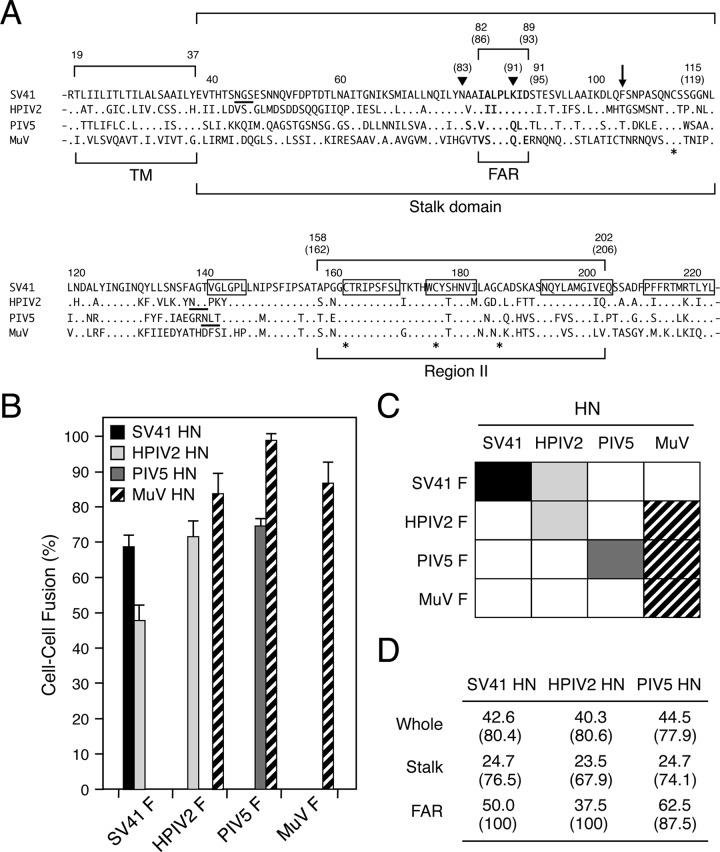
HN proteins of HPIV2 and MuV can trigger noncognate F proteins. (A) Amino acid sequence alignment of the rubulavirus HN proteins. The dots in the sequences indicate the amino acids that are identical to the counterparts of the SV41 HN protein, and the asterisks below the sequences denote the positions of cysteine residues that are conserved among all HN proteins. The positions of the PIV5 HN stalk domain (28) and HPIV2 HN region II (10) are indicated. The amino acid numbers of the SV41 HN protein are shown above the sequences; those of the HPIV2 HN protein are shown in parentheses. Filled triangles indicate the positions of N83 and K91 of the HPIV2 HN protein, which are the constituents of the MAb M1-1A epitope (10, 24). The downward arrow indicates the position of F104 of the SV41 HN protein; its counterparts of the other four HN proteins are conserved threonine residues. The amino acid regions that comprise the five β strands (β1) in the first blade of the 6-bladed propeller (32) are boxed. Consensus sequences for N-linked glycosylation are underlined. TM, transmembrane domain; FAR, F-activating region. (B) F-triggering activity of the rubulavirus HN proteins. Subconfluent BHK cells in six-well culture plates were transfected with 0.2 μg/well of HN expression vector together with 2 μg/well of cognate or noncognate F expression vector, except that 1 μg/well of expression vector was used for the PIV5 HN protein. After 24 h of incubation at 37°C, the average fusion index was determined as described in Materials and Methods; error bars indicate standard deviations. (C) Substitutability of the rubulavirus HN proteins. Events of functional HN-F interaction are indicated as filled or hatched boxes on the basis of the results presented in panel B. (D) Amino acid sequence identity and similarity among the rubulavirus HN proteins. Percent identity (or percent similarity) of the amino acid sequences between the MuV HN protein and the other HN proteins was calculated individually.