FIG 9.
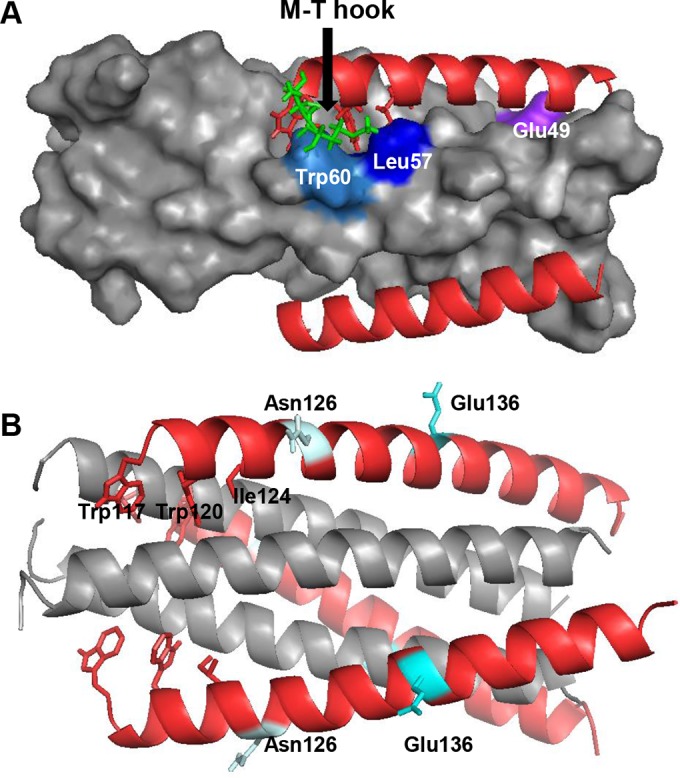
Modeling of MTSC22 resistance mutations on the 6-HB by the program PyMOL. (A) Analysis of the E49K and L57R mutations on the 6-HB formed by MTSC22 and an NHR-derived peptide (Protein Data Base ID code 3VU6). The pocket on the NHR helices is inserted by three hydrophobic residues (Trp117, Trp120, and Ile124) from the N-terminal pocket-binding domain of MTSC22. The M-T hook structure (green) of MTSC22 interacts with both Leu57 (blue) and Trp60 (marine) located at the left wall of the deep pocket. Therefore, Leu57 has extensive hydrophobic interactions with both the M-T hook residues and the pocket-binding residues to stabilize the binding of MTSC22. The residue Glu49 (purple) is located closely upstream of the pocket, and it can mediate a salt-bridge interaction with the positively charged lysine at position 20 (Lys20) of MTSC22. (B) Analysis of the N126K and E136G mutations on the 6-HB structure formed by N36 and C34 (Protein Data Base ID code 1AIK). The residues Asn126 and Glu136 are located, respectively, at the c and f positions of a CHR helix, corresponding to the outside of the binding surface of a CHR helix.
