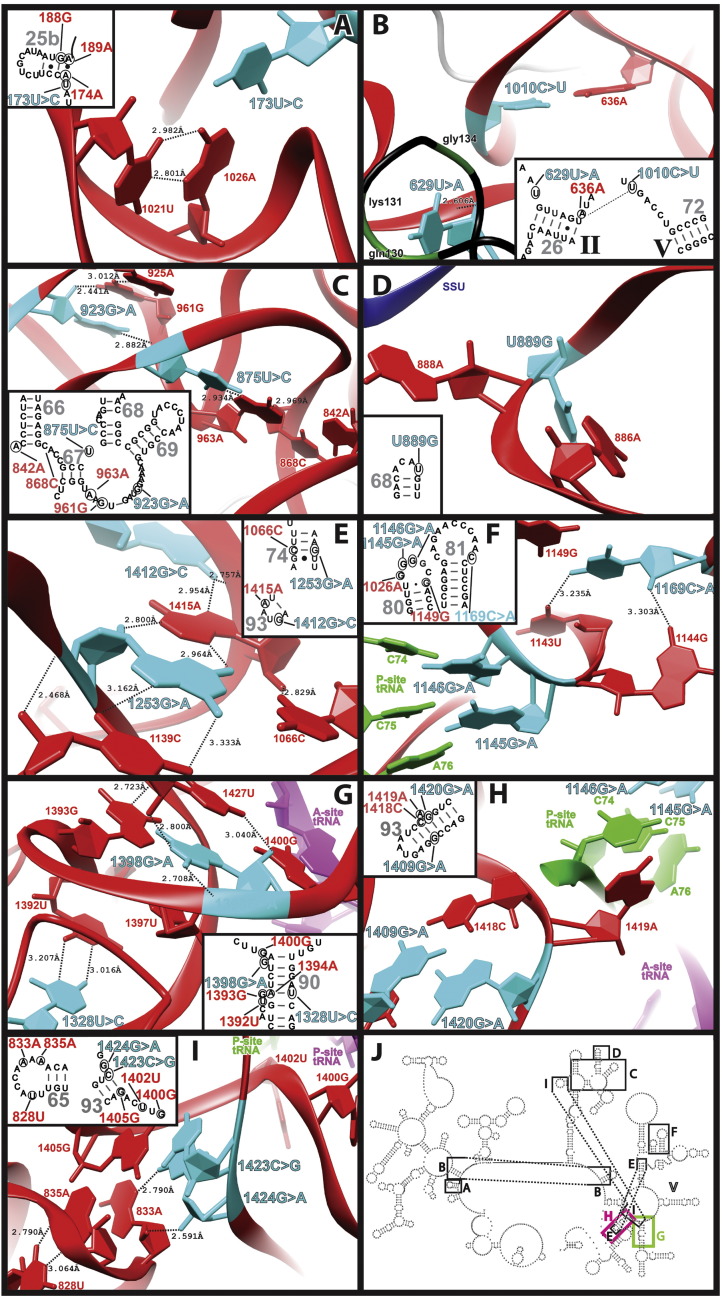
Fig. 2.
Structural analysis of variants with high disruptive potential. A) 173U > C (m.1843T > C). B) 629U > A (m.2299T > A) and 1010U > C (m.2680T > C). C) 875U > C (m.2545T > C) and 923G (m.2593G). D) 889U > G (m.2559T > G). E) 1253G > A (m.2923G > A) and 1412G > C (m.3082G > C). F) 1145G > A (m.2815G > A), 1146G > A (m.2816G > A), and 1169C > A (m.2839C > A). G) 1328U > C (m.2998T > C) and 1398G > A (m.3068G > A). H) 1420G > A (m.3090G > A) and 1409G > A (m.3079G > A). I) 1423C > G (m.3093C > G) and 1424G > A (m.3094G > A). J) Secondary-structure diagram with explanatory boxes indicating the location of the sites of mutation shown in panels A through I. Boxes G and H are coloured in magenta and light green to simplify the visualization of the figure. Variant sites in A–I are shown in aquamarine, associated sites in red. Ligands and atomic distances are indicated. tRNA structures in F, G, H, and I and SSU structure in D were superimposed as described in Methods. Insets correspond to the areas boxed in panel J. Helices are indicated in insets with large grey numbers. rRNA domains are indicated in insets with Roman numerals. Variant and associated sites in insets are colour coded as on main panel.