Abstract
A wealth of experimental data exists describing the elementary building blocks of complex physiological systems. However, it is increasingly apparent in the biomedical sciences that mechanisms of biological function cannot be observed or readily predicted via study of constituent elements alone. This is especially clear in the longstanding failures in prediction of effects of drug treatment for heart rhythm disturbances. These failures stem in part from classical assumptions that have been made in cardiac antiarrhythmic drug development – that a drug operates by one mechanism via one target receptor that arises from one gene.
Introduction
A challenge of the current research era is to integrate data in physiological networks to reveal emergent mechanisms of disease and to facilitate prediction and development of therapeutic interventions. Computational modeling and simulation constitute some of the most promising methodologies to reveal fundamental biological principles and mechanisms, model effects of interactions between system components and predict emergent disease processes and treatment effects. At present, there is no reasonable, efficient and cost-effective alternative experimental or clinical strategy that can achieve all of these goals. New computational methods are being developed that take advantage of high-performance computing technologies and strong interdisciplinary connections to experimentalists and clinicians to inform and verify the models 1. The resulting frameworks may ultimately be scaled up and automated for use in industry, academia and clinical medicine that can interact with other developing technologies including medium and high-throughput instrumentation (such as PatchExpress, IonWorks), synthetic drug development 2 and therapy by utilizing data from patient induced pluripotent stem (iPS) cell-derived cardiomyocytes 3.
Failure to predict drug effects
In the specific context of cardiac arrhythmia therapy, the vital hindrance to pharmacological treatment of patients with electrical rhythm disturbances is a long failed history in predicting effective or harmful action of drugs. For example, the CAST 4 and SWORD 5 clinical trials, showed that common antiarrhythmics increased mortality and risk of sudden cardiac death in post-infarction patients. Almost thirty years after CAST began, there is still no available approach to differentiate potentially useful and potentially harmful drugs for treating arrhythmia .
The first necessary step for developing computational models to predict drug effects on excitable cells is modeling the kinetics of primary and off-target drug and metabolite interactions with subcellular targets. The bulk of antiarrhythmics target ion channels which, in response to changes in voltage, undergo conformational changes that result in changes to apparent affinities of drugs for their receptors. These interactions have recently been modeled at the atomic scale in simulated docking and molecular dynamics (MD) simulations, as well as at the level of the channel function on the millisecond timescale of channel gating 6–7,8–10,11–14,15. Drug channel models have also been incorporated into cellular level computational models to test their effects on cell level parameters to search for antiarrhythmic or overt proarrhythmic potential 12–14, 16–18,19. Computational studies have also been carried out in tissue representations in one and two dimensions and even in high-resolution reconstructions of human virtual ventricles 14, 20,1,21–22. These computational studies have begun to improve understanding of antiarrhythmic drug actions across multiple spatial scales of the cardiac system, from molecule, to channel, to cell, to tissue, to heart.
Prediction of the structural determinants of drug interactions with discrete conformation states of ion channels in Rosetta
Structural modeling of ion channel interactions with drugs and other ligands is an important approach for current and future drug discovery efforts. Atomic scale modeling of drug receptor sites within an ion channel structure must be performed for multiple discrete conformational states to identify side chain and backbone atoms forming key drug-channel interactions. Such structural modeling of pore-forming and voltage-sensing domains of potassium and sodium channels has been successfully done using Rosetta-Membrane and Rosetta symmetry computational methods 23–25.
The process of modeling requires high-resolution structures of potassium and sodium channels that are used as templates for pairwise sequence alignments generated using the HHPred server26–28. The loops between transmembrane segments are modeled de novo if there are significant sequence differences between sequences of the ion channel of interest and the template ion channel structure. Several rounds of loop modeling28, 29 are performed and the lowest energy models are chosen as the best models for drug docking. Closed, open and inactivated states of ion channel pore-forming domain are generated based on available closed, open, and intermediate state templates from potassium and sodium channel pore-forming domain structures. Resting, intermediate, and activated states of the voltage-sensing domain are generated starting from available activated state templates from potassium and sodium channel voltage-sensing domain structures and new states are simulated using experimental data as constraints30–33.
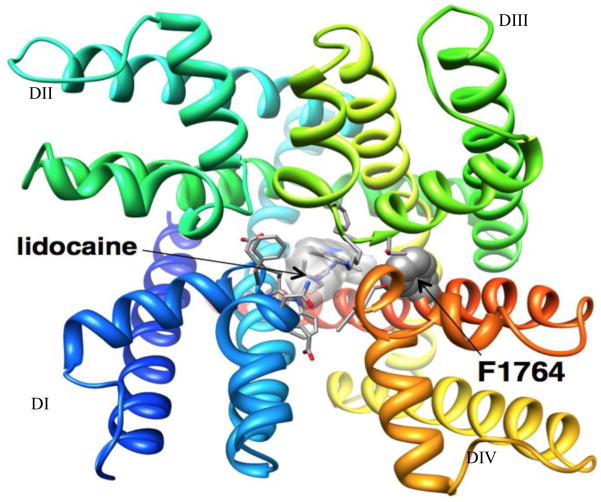
Ligand docking simulations are performed using the Rosetta-Ligand method6, 7 composed of three stages, which progress from low-resolution conformational sampling and scoring to full atom optimization using the all-atom energy function. The lowest energy structures from docking simulations are selected as the best models. Experimental data for mapping of key residues important for drug-channel interactions are used to evaluate accuracy of channel - drug models (see Figure 1). Structural modeling of drug-channel interactions at atomic scale may lead to design of novel high-affinity and subtype selective drugs for treatment of cardiac and neurological disorders.
Figure 1.
Docking of lidocaine in the Rosetta model of Nav1.5 channel pore. View of Nav1.5 - lidocaine model from the extracellular side of the membrane. Each domain is colored individually and labeled. Side chains of key residues for lidocaine binding are shown in space-filling and stick representation (Yarov-Yarovoy Laboratory).
Prediction of the drug affinity to discrete ion channel states with molecular dynamics simulations
MD simulation is an essential tool for studying biological systems, with recent increases in computer power allowing simulations of realistic ion channel systems on timescales relevant to function (e.g. 34). A promising avenue for drug discovery is to carry out MD simulations using homology (or de novo) models of mammalian ion channels (as described above) based on available high-resolution structures. In cases where drug activity is well conserved between mammallian and bacterial channels (e.g. Nav channels: 35,36), exploration of drug binding can be carried out directly using the structurally simpler bacterial channels. Figure 2 shows a typical Nav ion channel system studied using the DE Shaw Anton supercomputer for periods of many microseconds to reveal channel conduction, selectivity, conformational changes associated with activity, and drug accessibility and binding 37, 38. Such long timescale exploration of an ion channel can provide a new level of understanding, especially when combined with quantitative and predictive free energy methods.
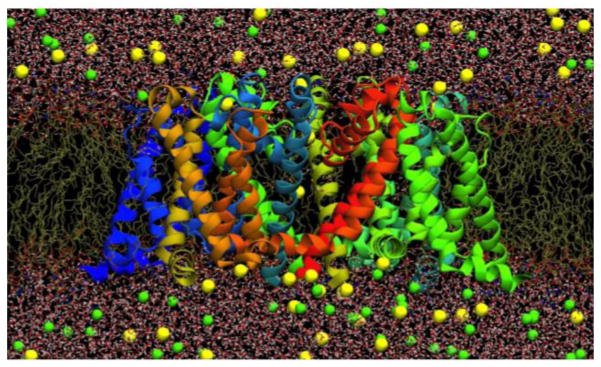
Figure 2.
Fully-atomistic model of the bacterial NavAb ion channel (~120,000 atoms; protein as ribbons, lipid tails as chains, water as sticks and ions as balls; Boiteux, C., I. Vorobyov and T. W. Allen. 2013. Submitted.).
Free energy calculations performed with fully-atomistic MD models provide the best means to quantify drug binding. Equilibrium sampling of the protein and ligand as a function of separation, using Umbrella Sampling 39, can be used to yield a free energy profile for drug binding. Here a sequence of independent simulations are carried out to ensure sampling along the reaction coordinate, which can then be optimally combined via the Weighted Histogram Analysis Method 40. One may instead use Free Energy Perturbation 41, where one alchemically decouples the ligand from its surroundings in the dissociated reference state, and then couples it to the surroundings in the binding pocket, to yield an absolute (or standard) binding free energy. Much effort has gone into achieving accurate results using staged protocols that avoid overlapping atomic centers when repulsive interactions are turned on and off 42, 43. An easier task is to transform one ligand into another to give a relative free energy of binding (e.g. to explore different functionalities or mutants).
Each of these methods faces inherent challenges due to the difficulties in sampling all configurations of the ligand and the protein as they approach, as well as the flexible response, which may involve rapid local, or global conformational changes that occur on exceedingly long timescales. Methods have been developed to overcome these challenges, such as “confine-and-release” (e.g. 44) or the “double decoupling method” (e.g. 45), where independent simulations restrain protein and ligand in conformation, position and/or orientation, to narrow the configurational space to be sampled, leading to improved statistical convergence 46 . Drug binding free energies are, however, ultimately reliant on the quality of the model. Today’s biomolecular force fields yield free energies that are in good agreement with experiments (e.g. 47), although require correct parameterization of drug compounds consistent with the protein and solvent force field (e.g. 48), and may still have limitations due to lacking explicit electronic polarizability, or unknown or changing protonation states 45. Such factors require careful attention for studies of drug binding that seek to be truly predictive and lead to improved rational ion channel drug design.
Kinetic models describing the interaction of drugs with cardiac ion channels
In order to accurately predict ion channel drug effects in higher dimensions, the intrinsic and explicit dynamical complexity of the drug kinetics is increasingly being considered in computational model representations (Figure 3). Early studies of drug effects on cardiac ion channels relied on pore-block models 20, which did not include the complex features of drug-channel kinetics that fundamentally emerge to alter cardiac rhythms in higher dimensions.
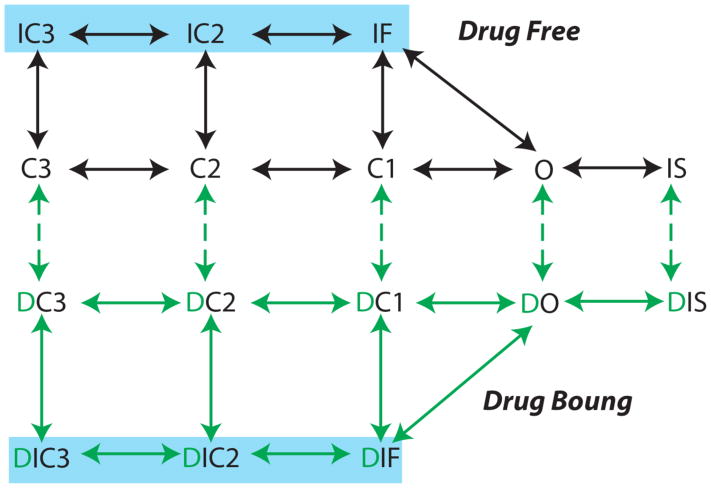
Figure 3.
An example for a Markov model representation of drug free and drug bound states for a model of the voltage gated Na channel (drug bound states are in green). In construction of a Markov based kinetic model representation of drug channel interactions, each drug free state in the model typically has a corresponding drug associated state. This assumption derives from the modulated receptor hypothesis, where any channel state can be drug-bound, although drugs exhibit distinct affinities for discrete states 49. Measured affinities and drug diffusion rates from experiments are used to constrain the drug “on” and “off” rates in the model. Measured diffusion rates (D) are used to inform drug on rates “kon” = [drug] * D and affinities (Kd) to discrete conformations that determine drug off rates “koff” = Kd * D. Model rate constants are then further constrained by optimization to data and microscopic reversibility 50.
Predictive models of drugs on the cellular level action potential
Because drugs alter the action potential waveform, which in turn affects the potency of drugs, the strong bi-directional feedback loop between cellular level electrical activity and drug potency has been studies using modeling and simulation approaches 51. The kinetic interactions of drugs with ion channels are modified by cellular action potential properties including morphology, duration and frequency. Computational modeling allows for investigation of drug effects on action potentials (APs) over a wide range of therapeutically relevant ranges of drug concentration and clinically relevant pacing frequencies (typically 60 – 220 beats per minute (BPM)). Arrhythmia susceptibility parameters can be tracked over the course of a simulation, including cell excitability (maximum upstroke velocity of the AP (V/s)), action potential duration (APD), cell refractoriness and APD restitution as described previously 52.
An additional complicating factor is the natural inter-subject variability in the electrophysiological activity of cardiac cells both within and between individuals. This variability has also been modeled, which should allow prediction of the response of specific cells (and cell populations) from specific hearts to disease and therapies 53,18,54.
Although cellular level studies can plausibly suggest reduced or increased arrhythmia vulnerability, reentrant arrhythmias are fundamentally an emergent property of the cardiac system that can only be observed and studied in tissue. Thus, models have been developed to predict drug effects in higher dimensions that include spatial dimension and cellular coupling.
Predictive models of drug effects on cardiac tissue
In cardiac tissue, electrotonic coupling leads to unpredictable emergent responses to drug application. An example is in a recent study, where mild depression of single cell cellular excitability was predicted in response to flecainide, suggesting its therapeutic potential to suppress ectopic arrhythmia triggers 55. No overt proarrhythmic potential was ever observed in cells. In tissue level simulations, the outcome was dramatically different. Substantial use-dependent block with flecainide resulted in insufficient Na channel availability for successful conduction, a higher dimensional phenomenon that emerged as a result of increased electrotonic load in coupled tissue. Proarrhythmic conduction block sometimes led to development of tachycardia indicated by spiral wave reentry, verified experimentally in rabbit heart and in MRI-based human 3D ventricle models 55.
The general approach to predict effects of antiarrhythmic drugs and metabolites in ventricular cells and tissues is: (1) Drug-channel models will be incorporated into human computational ventricular cell and tissue models. (2) Simulations are run and the arrhythmia vulnerability parameters: APD restitution, Ca2+ dynamics, dispersion of repolarization and reentry wavelength can be tracked. (3) Sensitivity analysis is performed. (4) When possible simulations may be validated experimentally.
One-dimensional (1D) simulations
1D simulations are useful to coarsely identify parameter regimes of interest with a computationally tractable model. Regimes exhibiting compelling dynamics can then be investigated in higher dimensions. This improves efficiency and ensures that higher dimensional computationally expensive simulations will be constrained. In one dimension, the following parameters can be readily tracked in the simulation: Conduction velocity (CV): Antiarrhythmic drugs can reduce Na current by directly inhibiting Na channels, or by increasing APD sufficiently so that Na channels have insufficient recovery time during diastolic intervals. CV depression can cause conduction block and reentry. CV is typically calculated between two cells at the time of the maximum dV/dt on the upstroke of the action potential. Drug concentration for conduction block (CB): Conduction block is potentially proarrhythmic because it can cause dispersion of repolarization; if unidirectional block occurs, reentrant arrhythmias can ensue. CB can precipitate wavebreak in the transition to fibrillation 56. Thus, simulations of conduction block over a range of physiological frequencies and drug concentrations can be simulated with escalating drug concentrations can be applied until block occurs. Calculation of the vulnerable window to unidirectional conduction block: It has been long known that a period of vulnerability exists whereby electrical stimulation can initiate self-sustaining spiral waves 57,58 capable of degeneration into fibrillatory rhythms. Starmer et al. 59–61, developed an approach to systematically determine the likelihood of arrhythmia induced by spontaneous ventricular stimuli with drugs. One-dimensional tissue simulations are useful to assess the “vulnerable window” to unidirectional block and retrograde conduction, which suggests reentrant arrhythmia in higher dimensions 61, 60. The procedure is described in 55. Quantification of Arrhythmia Probability: The vulnerable window (VW) is not sufficient to predict arrhythmia risk - the refractory period must also be considered. The Starmer metric was developed to quantify arrhythmia risk and can be applied in the absence or presence of simulated drugs 59.
Pause-induced or short-long-short arrhythmia trigger
Torsades de pointes arrhythmias my be triggered by specific pacing anomalies including pauses or short-long-short pacing sequences – especially in the setting of acquired long-QT syndrome resulting from IKr block 62. A sinus pause sets the stage for exaggerated heterogeneous action potential prolongation in response to the subsequent stimulation. Such prolongation may even include early afterdepolarizations (EAD). This can lead to triggered ectopy and set the stage for the clinically observed “cascade effect” leading to torsades de pointes. Simulations can allow for tracking the extent of APD prolongation following a pause and/or EAD development after a pause following steady state pacing 63–65–13, 66–68.
Two-dimensional (2D) simulations
2D simulations can be undertaken to predict if proarrhythmic phenomena observed in lower dimensions cause reentrant arrhythmias and/or spiral wave breakup. The change in voltage in space and time are tracked in the simulation 69. An example of model prediction of drug effects from our group shows 2D stable reentry induced by flecainide after static pacing (S1) followed by an S2 within the vulnerable window (computed in 1-dimension for efficiency) is shown in Figure 3. Arrhythmia vulnerability parameters as described for one-dimensional tissue can be tracked in two dimensions 55,70,71,72. In two dimensions, reentry wavelength and period can also be tracked to investigate head-tail interactions.
Simulation in high-resolution image based reconstructed geometrically realistic human ventricular models
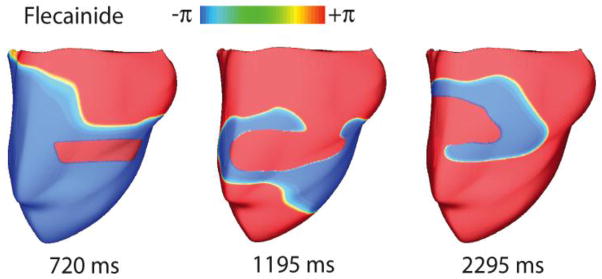
High-dimensional drug simulations have been recently undertaken as computational resources are increasingly accessible 1, 20,21–22. The Trayanova lab recently debuted simulations of drug effects in multiple MRI-based anatomically detailed 3-dimensional models of the human ventricles. An example is shown for flecainide-induced reentry in Figure 4. 3D ventricles were paced from the apex at a rate of 120 BPM with high dose flecainide (2 μM). An ectopic stimulus inside the vulnerable window (phase maps are shown) approximately halfway up the ventricles initiated a persistent figure-of-eight reentrant wave with flecainide, demonstrated the proarrhythmic potential off flecainide with an ectopic stimulus applied with the vulnerable window.
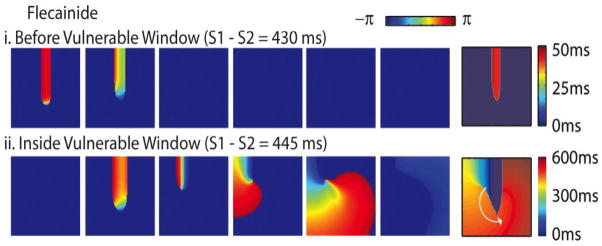
Figure 4.
Flecainide in a 2D tissue model. Phase maps for flecainide (2 μM) (scale on top: red is wavefront, and blue is repolarized (though not necessarily recovered from drug block)). Right panels are activation isochrones. A premature impulse applied in the wake of the preceding wave (i) before or (ii) in the vulnerable window (from 14.
Future directions for modeling and simulation in pharmacology: Why do promising drugs fail?
Computational approaches can and should be used to probe the mechanisms of drug failures 73. This is an area of vital importance, since understanding the mechanisms drug failure are essential to rule out compounds in early preclinical drug screening. Sequential component dissection can be used in model frameworks to reveal the mechanisms of drug failure. A recent investigation focused on the mechanisms of failure of the once promising antiarrhythmic drug flecainide, the subject of the cardiac arrhythmia suppression trial (CAST), which startlingly showed increased mortality with flecainide over placebo. We used a computational process to first confirm experimental findings: no overt proarrhythmic potential was ever observed in cells55. In tissue level simulations, the outcome was dramatically different. Substantial use-dependent block by flecainide (an intrinsic dynamical property of channel block) resulted in failed impulse conduction, a higher dimensional phenomenon that emerged as a result of increased electrotonic load in coupled tissue. Proarrhythmic conduction block led to development of tachycardia indicated by spiral wave reentry, which we verified experimentally 55. This emergent phenomenon was linked back to the fundamental mechanism - the drug kinetics of unblock, identified as the basic mechanism of failure. Computationally based models of cells and tissues can be constructed to predict emergent drug effects on cellular and tissue electrical activity, and then be deconstructed via component dissection to reveal mechanisms.
Conclusions
Computational pharmacology models are being developed that may serve as prototypes that are expandable, scalable and have potential for automation so that they can ultimately interact with other developing technologies including high throughput measurements (such as PatchExpress 74–79, Ionworks 80–85), drug design via new developments in synthetic biology 2, and even personalized medicine via drug screening in patients’ own induced pluripotent stem (iPS) cell-derived cardiomyocytes 3. All of these developing technologies are innovative and fantastic, but they focus only on constituent elements of the system. They can’t each alone solve the fundamental problem – that the effects of multifaceted drug interactions are emergent. But, these technologies in conjunction with the computational based methods under development to predict emergent effects of drugs on excitable rhythms may form an interactive technology driven process that can be used in industry for drug and disease screening, in academia for research and development and in the clinic for patient oriented medicine.
Figure 5.
Reentry in 3D models of the human ventricle from the Trayanova Lab 14. (A) Phase maps of a sustained figure-of-eight reentry with 2 μM flecainide at 120 BPM in response to an S2 within the vulnerable window.
Footnotes
Conflict of interest
The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zemzemi N, Bernabeu MO, Saiz J, Cooper J, Pathmanathan P, Mirams GR, Pitt-Francis J, Rodriguez B. Computational assessment of drug-induced effects on the electrocardiogram: From ion channel to body surface potentials. Br J Pharmacol. 2013;168:718–733. doi: 10.1111/j.1476-5381.2012.02200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nattel S, Carlsson L. Innovative approaches to anti-arrhythmic drug therapy. Nature Reviews Drug Discovery. 2006;5:1034–1049. doi: 10.1038/nrd2112. [DOI] [PubMed] [Google Scholar]
- 3.Braam SR, Tertoolen L, van de Stolpe A, Meyer T, Passier R, Mummery CL. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010;4:107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Preliminary report: Effect of encainide and flecainide on mortality in a randomized trial of arrhythmia suppression after myocardial infarction. The cardiac arrhythmia suppression trial (cast) investigators. N Engl J Med. 1989;321:406–412. doi: 10.1056/NEJM198908103210629. [DOI] [PubMed] [Google Scholar]
- 5.Waldo AL, Camm AJ, deRuyter H, Friedman PL, MacNeil DJ, Pauls JF, Pitt B, Pratt CM, Schwartz PJ, Veltri EP. Effect of d-sotalol on mortality in patients with left ventricular dysfunction after recent and remote myocardial infarction. The sword investigators. Survival with oral d-sotalol. Lancet. 1996;348:7–12. doi: 10.1016/s0140-6736(96)02149-6. [DOI] [PubMed] [Google Scholar]
- 6.Meiler J, Baker D. Rosettaligand: Protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 7.Davis IW, Baker D. Rosettaligand docking with full ligand and receptor flexibility. Journal of molecular biology. 2009;385:381–392. doi: 10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Zhang JZ, Yarov-Yarovoy V, Scheuer T, Karbat I, Cohen L, Gordon D, Gurevitz M, Catterall WA. Structure-function map of the receptor site for beta-scorpion toxins in domain ii of voltage-gated sodium channels. The Journal of biological chemistry. 2011;286:33641–33651. doi: 10.1074/jbc.M111.282509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang JZ, Yarov-Yarovoy V, Scheuer T, Karbat I, Cohen L, Gordon D, Gurevitz M, Catterall WA. Mapping the interaction site for a beta-scorpion toxin in the pore module of domain iii of voltage-gated na(+) channels. The Journal of biological chemistry. 2012;287:30719–30728. doi: 10.1074/jbc.M112.370742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Q, Bett GC, Rasmusson RL. Markov models of use-dependence and reverse use-dependence during the mouse cardiac action potential. PloS one. 2012;7:e42295. doi: 10.1371/journal.pone.0042295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malisi C, Schumann M, Toussaint NC, Kageyama J, Kohlbacher O, Hocker B. Binding pocket optimization by computational protein design. PloS one. 2012;7:e52505. doi: 10.1371/journal.pone.0052505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clancy CE, Zhu ZI, Rudy Y. Pharmacogenetics and anti-arrhythmic drug therapy: A theoretical investigation. American journal of physiology. Heart and circulatory physiology. 2007;292:H66–75. doi: 10.1152/ajpheart.00312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno JD, Yang PC, Bankston JR, Grandi E, Bers DM, Kass RS, Clancy CE. Ranolazine for congenital and acquired late ina linked arrhythmias: In silico pharmacologic screening. Circulation research. 2013 doi: 10.1161/CIRCRESAHA.113.301971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno JD, Zhu ZI, Yang PC, Bankston JR, Jeng MT, Kang C, Wang L, Bayer JD, Christini DJ, Trayanova NA, Ripplinger CM, Kass RS, Clancy CE. A computational model to predict the effects of class i anti-arrhythmic drugs on ventricular rhythms. Science translational medicine. 2011;3:98ra83. doi: 10.1126/scitranslmed.3002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carpenter TS, Lau EY, Lightstone FC. Identification of a possible secondary picrotoxin-binding site on the gabaa receptor. Chemical research in toxicology. 2013;26:1444–1454. doi: 10.1021/tx400167b. [DOI] [PubMed] [Google Scholar]
- 16.Sarkar AX, Sobie EA. Quantification of repolarization reserve to understand interpatient variability in the response to proarrhythmic drugs: A computational analysis. Heart rhythm : the official journal of the Heart Rhythm Society. 2011;8:1749–1755. doi: 10.1016/j.hrthm.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Veroli GY, Davies MR, Zhang H, Abi-Gerges N, Boyett MR. High-throughput screening of drug-binding dynamics to herg improves early drug safety assessment. American journal of physiology. Heart and circulatory physiology. 2013;304:H104–117. doi: 10.1152/ajpheart.00511.2012. [DOI] [PubMed] [Google Scholar]
- 18.Britton OJ, Bueno-Orovio A, Van Ammel K, Lu HR, Towart R, Gallacher DJ, Rodriguez B. Experimentally calibrated population of models predicts and explains intersubject variability in cardiac cellular electrophysiology. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2098–2105. doi: 10.1073/pnas.1304382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Recanatini M, Cavalli A, Masetti M. Modeling herg and its interactions with drugs: Recent advances in light of current potassium channel simulations. ChemMedChem. 2008;3:523–535. doi: 10.1002/cmdc.200700264. [DOI] [PubMed] [Google Scholar]
- 20.Brennan T, Fink M, Rodriguez B. Multiscale modelling of drug-induced effects on cardiac electrophysiological activity. Eur J Pharm Sci. 2009;36:62–77. doi: 10.1016/j.ejps.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Dux-Santoy L, Sebastian R, Felix-Rodriguez J, Ferrero JM, Saiz J. Interaction of specialized cardiac conduction system with antiarrhythmic drugs: A simulation study. IEEE transactions on bio-medical engineering. 2011;58:3475–3478. doi: 10.1109/TBME.2011.2165213. [DOI] [PubMed] [Google Scholar]
- 22.Obiol-Pardo C, Gomis-Tena J, Sanz F, Saiz J, Pastor M. A multiscale simulation system for the prediction of drug-induced cardiotoxicity. Journal of chemical information and modeling. 2011;51:483–492. doi: 10.1021/ci100423z. [DOI] [PubMed] [Google Scholar]
- 23.Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barth P, Schonbrun J, Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andre I, Bradley P, Wang C, Baker D. Prediction of the structure of symmetrical protein assemblies. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17656–17661. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soding J. Protein homology detection by hmm-hmm comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. Journal of molecular biology. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 28.Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. Journal of molecular biology. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 30.Yarov-Yarovoy V, Decaen PG, Westenbroek RE, Pan CY, Scheuer T, Baker D, Catterall WA. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E93–E102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decaen PG, Yarov-Yarovoy V, Scheuer T, Catterall WA. Gating charge interactions with the s1 segment during activation of a na+ channel voltage sensor. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18825–18830. doi: 10.1073/pnas.1116449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22498–22503. doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15142–15147. doi: 10.1073/pnas.0806486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen MO, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 35.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, Goodchild SJ, Ahern CA. Local anesthetic inhibition of a bacterial sodium channel. The Journal of general physiology. 2012;139:507–516. doi: 10.1085/jgp.201210779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boiteux C, Vorobyov I, Allen TW. Biophysical Journal. 2013;104:137a. [Google Scholar]
- 38.Boiteux C, Vorobyov I, Allen TW. 2013 submitted. [Google Scholar]
- 39.Torrie GM, Valleau JP. Non-physical sampling distributions in monte-carlo free-energy estimation - umbrella sampling. J Comput Phys. 1977;23:187–199. [Google Scholar]
- 40.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules .1. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 41.Kirkwood JG. Statistical mechanics of fluid mixtures. J Chem Phys. 1935;3:300–313. [Google Scholar]
- 42.Deng YQ, Roux B. Hydration of amino acid side chains: Nonpolar and electrostatic contributions calculated from staged molecular dynamics free energy simulations with explicit water molecules. Journal of Physical Chemistry B. 2004;108:16567–16576. [Google Scholar]
- 43.Weeks JD, Chandler D, Andersen HC. Role of repulsive forces in determining equilibrium structure of simple liquids. J Chem Phys. 1971;54:5237. [Google Scholar]
- 44.Mobley DL, Chodera JD, Dill KA. Confine-and-release method: Obtaining correct binding free energies in the presence of protein conformational change. J Chem Theory Comput. 2007;3:1231–1235. doi: 10.1021/ct700032n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng YQ, Roux B. Computations of standard binding free energies with molecular dynamics simulations. Journal of Physical Chemistry B. 2009;113:2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gumbart JC, Roux B, Chipot C. Efficient determination of protein-protein standard binding free energies from first principles. J Chem Theory Comput. 2013;9:3789–3798. doi: 10.1021/ct400273t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mobley DL, Bayly CI, Cooper MD, Shirts MR, Dill KA. Small molecule hydration free energies in explicit solvent: An extensive test of fixed-charge atomistic simulations. J Chem Theory Comput. 2009;5:350–358. doi: 10.1021/ct800409d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, MacKerell AD. Charmm general force field: A force field for drug-like molecules compatible with the charmm all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hille B. Local-anesthetics - hydrophilic and hydrophobic pathways for drug-receptor reaction. Journal of General Physiology. 1977;69:497–515. doi: 10.1085/jgp.69.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colquhoun D, Dowsland KA, Beato M, Plested AJ. How to impose microscopic reversibility in complex reaction mechanisms. Biophysical journal. 2004;86:3510–3518. doi: 10.1529/biophysj.103.038679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trenor B, Gomis-Tena J, Cardona K, Romero L, Rajamani S, Belardinelli L, Giles WR, Saiz J. In silico assessment of drug safety in human heart applied to late sodium current blockers. Channels (Austin) 2013;7 doi: 10.4161/chan.24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldhaber JI, Qu ZL, Garfinkel A, Duong T, Weiss JN. Determinants of action potential duration restitution in isolated ventricular myocytes. Circulation. 1997;96:3756–3756. [Google Scholar]
- 53.Yang PC, Clancy CE. In silico prediction of sex-based differences in human susceptibility to cardiac ventricular tachyarrhythmias. Front Physiol. 2012;3:360. doi: 10.3389/fphys.2012.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahrens-Nicklas RC, Clancy CE, Christini DJ. Re-evaluating the efficacy of beta-adrenergic agonists and antagonists in long qt-3 syndrome through computational modelling. Cardiovascular research. 2009;82:439–447. doi: 10.1093/cvr/cvp083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moreno JD, Zhu ZI, Yang PC, Bankston JR, Jeng MT, Kang C, Wang L, Bayer JD, Christini DJ, Trayanova NA, Ripplinger CM, Kass RS, Clancy CE. A computational model to predict the effects of class i anti-arrhythmic drugs on ventricular rhythms. Science Translational Medicine. 2011;3:98ra83. doi: 10.1126/scitranslmed.3002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiss JN, Qu Z, Chen P-S, Lin S-F, Karagueuzian HS, Hayashi H, Garfinkel A, Karma A. The dynamics of cardiac fibrillation. Circulation. 2005;112:1232–1240. doi: 10.1161/CIRCULATIONAHA.104.529545. [DOI] [PubMed] [Google Scholar]
- 57.Mines G. On circulating excitations in heart muscles and their possible relation to tachycardia and fibrillation. Trans Roy Soc Can. 1914:43–53. [Google Scholar]
- 58.Allessie MA, Bonke FI, Schopman FJ. Circus movement in rabbit atrial muscle as a mechanism of trachycardia. Circulation research. 1973;33:54–62. [PubMed] [Google Scholar]
- 59.Starmer CF. How antiarrhythmic drugs increase the rate of sudden cardiac death. International Journal of Bifurcation and Chaos. 2002;12:1953–1968. [Google Scholar]
- 60.Starmer CF, Biktashev VN, Romashko DN, Stepanov MR, Makarova ON, Krinsky VI. Vulnerability in an excitable medium - analytical and numerical-studies of initiating unidirectional propagation. Biophysical journal. 1993;65:1775–1787. doi: 10.1016/S0006-3495(93)81233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Starmer CF, Lastra aa, Nesterenko VV, Grant aO. Proarrhythmic response to sodium-channel blockade - theoretical-model and numerical experiments. Circulation. 1991;84:1364–1377. doi: 10.1161/01.cir.84.3.1364. [DOI] [PubMed] [Google Scholar]
- 62.Roden DM, Anderson ME. The pause that refreshes, or does it? Mechanisms in torsades de pointes. Heart. 2000;84:235–237. doi: 10.1136/heart.84.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo CH, Rudy Y. A dynamic model of the cardiac ventricular action potential. Ii. Afterdepolarizations, triggered activity, and potentiation. Circulation research. 1994;74:1097–1113. doi: 10.1161/01.res.74.6.1097. [DOI] [PubMed] [Google Scholar]
- 64.Viswanathan PC, Rudy Y. Cellular arrhythmogenic effects of congenital and acquired long-qt syndrome in the heterogeneous myocardium. Circulation. 2000;101:1192–1198. doi: 10.1161/01.cir.101.10.1192. [DOI] [PubMed] [Google Scholar]
- 65.Viswanathan PC, Rudy Y. Pause induced early afterdepolarizations in the long qt syndrome: A simulation study. Cardiovascular research. 1999;42:530–542. doi: 10.1016/s0008-6363(99)00035-8. [DOI] [PubMed] [Google Scholar]
- 66.Clancy CE, Rudy Y. Na(+) channel mutation that causes both brugada and long-qt syndrome phenotypes: A simulation study of mechanism. Circulation. 2002;105:1208–1213. doi: 10.1161/hc1002.105183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nuyens D, Stengl M, Dugarmaa S, Rossenbacker T, Compernolle V, Rudy Y, Smits JF, Flameng W, Clancy CE, Moons L, Vos MA, Dewerchin M, Benndorf K, Collen D, Carmeliet E, Carmeliet P. Abrupt rate accelerations or premature beats cause life-threatening arrhythmias in mice with long-qt3 syndrome. Nature medicine. 2001;7:1021–1027. doi: 10.1038/nm0901-1021. [DOI] [PubMed] [Google Scholar]
- 68.Clancy CE, Rudy Y. Cellular consequences of herg mutations in the long qt syndrome: Precursors to sudden cardiac death. Cardiovascular research. 2001;50:301–313. doi: 10.1016/s0008-6363(00)00293-5. [DOI] [PubMed] [Google Scholar]
- 69.ten Tusscher KHWJ, Panfilov AV. Alternans and spiral breakup in a human ventricular tissue model. American journal of physiology. Heart and circulatory physiology. 2006;291:H1088–1100. doi: 10.1152/ajpheart.00109.2006. [DOI] [PubMed] [Google Scholar]
- 70.Zhu ZI, Clancy CE. L-type ca2+ channel mutations and t-wave alternans: A model study. American journal of physiology. Heart and circulatory physiology. 2007;293:H3480–3489. doi: 10.1152/ajpheart.00476.2007. [DOI] [PubMed] [Google Scholar]
- 71.Yang PC, Kurokawa J, Furukawa T, Clancy CE. Acute effects of sex steroid hormones on susceptibility to cardiac arrhythmias: A simulation study. PLoS computational biology. 2010;6:e1000658. doi: 10.1371/journal.pcbi.1000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakamura H, Kurokawa J, Bai C-X, Asada K, Xu J, Oren RV, Zhu ZI, Clancy CE, Isobe M, Furukawa T. Progesterone regulates cardiac repolarization through a nongenomic pathway: An in vitro patch-clamp and computational modeling study. Circulation. 2007;116:2913–2922. doi: 10.1161/CIRCULATIONAHA.107.702407. [DOI] [PubMed] [Google Scholar]
- 73.Sorger PK, Allerheiligen SRB. Quantitative and systems pharmacology in the postgenomic era: New approaches to discovering drugs and understanding therapeutic mechanisms. Tech Rep. 2011 [Google Scholar]
- 74.Penniman JR, Kim DC, Salata JJ, Imredy JP. Assessing use-dependent inhibition of the cardiac na(+/−) current (i(na)) in the patchxpress automated patch clamp. Journal of pharmacological and toxicological methods. 2010;62:107–118. doi: 10.1016/j.vascn.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 75.Mo ZL, Faxel T, Yang YS, Gallavan R, Messing D, Bahinski A. Effect of compound plate composition on measurement of herg current ic(50) using patchxpress. Journal of pharmacological and toxicological methods. 2009;60:39–44. doi: 10.1016/j.vascn.2009.04.198. [DOI] [PubMed] [Google Scholar]
- 76.Zeng H, Penniman JR, Kinose F, Kim D, Trepakova ES, Malik MG, Dech SJ, Balasubramanian B, Salata JJ. Improved throughput of patchxpress herg assay using intracellular potassium fluoride. Assay Drug Dev Technol. 2008;6:235–241. doi: 10.1089/adt.2007.116. [DOI] [PubMed] [Google Scholar]
- 77.Trepakova ES, Malik MG, Imredy JP, Penniman JR, Dech SJ, Salata JJ. Application of patchxpress planar patch clamp technology to the screening of new drug candidates for cardiac kcnq1/kcne1 (i ks) activity. Assay Drug Dev Technol. 2007;5:617–627. doi: 10.1089/adt.2007.091. [DOI] [PubMed] [Google Scholar]
- 78.Ly JQ, Shyy G, Misner DL. Assessing herg channel inhibition using patchxpress. Clin Lab Med. 2007;27:201–208. doi: 10.1016/j.cll.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 79.Dubin AE, Nasser N, Rohrbacher J, Hermans AN, Marrannes R, Grantham C, Van Rossem K, Cik M, Chaplan SR, Gallacher D, Xu J, Guia A, Byrne NG, Mathes C. Identifying modulators of herg channel activity using the patchxpress planar patch clamp. J Biomol Screen. 2005;10:168–181. doi: 10.1177/1087057104272295. [DOI] [PubMed] [Google Scholar]
- 80.Bridal TR, Margulis M, Wang X, Donio M, Sorota S. Comparison of human ether-a-go-go related gene screening assays based on ionworks quattro and thallium flux. Assay Drug Dev Technol. 2010;8:755–765. doi: 10.1089/adt.2010.0267. [DOI] [PubMed] [Google Scholar]
- 81.Jow F, Shen R, Chanda P, Tseng E, Zhang H, Kennedy J, Dunlop J, Bowlby MR. Validation of a medium-throughput electrophysiological assay for kcnq2/3 channel enhancers using ionworks ht. J Biomol Screen. 2007;12:1059–1067. doi: 10.1177/1087057107307448. [DOI] [PubMed] [Google Scholar]
- 82.Harmer AR, Abi-Gerges N, Easter A, Woods A, Lawrence CL, Small BG, Valentin JP, Pollard CE. Optimisation and validation of a medium-throughput electrophysiology-based hnav1.5 assay using ionworks. Journal of pharmacological and toxicological methods. 2008;57:30–41. doi: 10.1016/j.vascn.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 83.Bridgland-Taylor MH, Hargreaves AC, Easter A, Orme A, Henthorn DC, Ding M, Davis AM, Small BG, Heapy CG, Abi-Gerges N, Persson F, Jacobson I, Sullivan M, Albertson N, Hammond TG, Sullivan E, Valentin JP, Pollard CE. Optimisation and validation of a medium-throughput electrophysiology-based herg assay using ionworks ht. Journal of pharmacological and toxicological methods. 2006;54:189–199. doi: 10.1016/j.vascn.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 84.Sorota S, Zhang XS, Margulis M, Tucker K, Priestley T. Characterization of a herg screen using the ionworks ht: Comparison to a herg rubidium efflux screen. Assay Drug Dev Technol. 2005;3:47–57. doi: 10.1089/adt.2005.3.47. [DOI] [PubMed] [Google Scholar]
- 85.Schroeder K, Neagle B, Trezise DJ, Worley J. Ionworks ht: A new high-throughput electrophysiology measurement platform. J Biomol Screen. 2003;8:50–64. doi: 10.1177/1087057102239667. [DOI] [PubMed] [Google Scholar]