Abstract
Glioblastoma is a type of glioma with a relatively higher degree of malignancy that may result in severe intracranial hypertension and focal symptoms. Surgery is the preferred treatment modality. Combination therapy including radiotherapy, chemotherapy, gene therapy, immunotherapy and targeted therapy have also been employed. However, due to the invasiveness and pathogenesis of the disease, such treatments do not yield satisfactory outcomes. The aim of the present study was to examine the expression of microRNA (miR)-184 in Janus kinase 2/signal transducer and activator of transcription 3 (JAK2/STAT3) signaling pathway in the mechanism of glioblastoma formation, thus providing a new basis for the mechanism of glioblastoma induction. The LN18 cell line was employed in the present study. After undergoing thawing, culturing and passaging processes, the cells were divided into the set control group, miR-184 mimic group (transfer miR-184 simulator) and miR-184 group. The expression of miR-184 was detected using quantitative polymerase chain reaction. An MTT assay was used to detect the proliferation ability of glioma cells, and clone formation ability was also detected. The cell scratch and invasion assays were used to identify the cell invasion ability. Western blotting was performed to detect the expression level of p-JAK2 and p-STAT3 proteins. The results showed that compared to the control group, the expression of miR-184 in the miR-184 mimic group increased. Cell proliferation, as well as clone formation and invasion ability were enhanced. The number of cells penetrating septum, as well as the expression of p-JAK2 and p-STAT3 proteins were increased. Differences were statistically significant (P<0.05). By contrast, compared to the control group, the expression of miR-184 in the miR-184 inhibitory group decreased. Cell proliferation, as well as clone formation and invasion ability were reduced. The number of cells penetrating septum, as well as the expression of p-JAK2 and p-STAT3 proteins were reduced. Differences were statistically significant (P<0.05). In conclusion, the results of the present study have shown that miR-184 may be involved in the formation of glioblastoma and influence the expression of JAK2/STAT3 signaling pathway.
Keywords: microRNA-184, glioblastoma, Janus kinase 2/signal transducer and activator of transcription 3 signal pathway, mechanism
Introduction
Glioma is a high-grade malignant primary intracranial tumor, with a high morbidity and mortality rate (1). Glioblastoma is a type of glioma characterized by a relatively high degree of malignancy. Early-stage gliobastoma may lead to severe intracranial hypertension, and focal symptoms and signs (1). Surgery constitutes the most commonly used treatment modality in early-stage glioblastoma (1). Combined therapy including radiotherapy, chemotherapy, gene therapy, immunotherapy and targeted therapy are employed in later-stage glioblastoma (1). However, the clinical effects of these therapies and patient prognosis are far from ideal due to the unmanageable invasiveness and unclear pathogenesis of the disease. It has been found that the expression of microRNA (miR) is closely associated with the formation of glioma (2,3). Janus kinase 2/signal transducer and activator of transcription 3 (JAK2/STAT3) signaling pathways are important processes for the formation and transmission of tumors (4).
The aim of the present study was to examine the expression of miR-184 in JAK2/STAT3 signaling pathways in the formation of glioblastoma to provide a new basis for the development of the mechanism of glioblastoma.
Materials and methods
Materials
The LN28 glioblastoma cell line was purchased from the Chinese Academy of Sciences Cell Bank. The following experimental instruments were used: Micropipette tip (Rainin Instrument LLC, Oakland, CA, USA), optical microscope (Olympus Corp., Tokyo, Japan), polymerase chain reaction (PCR) TC-XP (Bioer Technology Co., Ltd., Hangzhou, China), constant temperature incubator (Changzhou Huapuda Instrument Co. Ltd., Changzhou, China), paraffin slicing machine (Leica, Mannheim, Germany) and tissue embedder (Leica). TRIzol reagent, RNase-free, reverse transcription (RT)-PCR primers, RT kit, quantitative PCR (qPCR) kit, Express SYBR-GreenER miR qPCR kits, NCode VILO miR cDNA (Invitrogen Life Technologies, Carlsbad, CA, USA) and the miR extraction kit (Roche Diagnostics GmbH, Mannheim, Germany) were employed. The rabbit polyclonal primary antibodies p-JAK2 (Cat. no. GTX101132, 1:500) and p-STAT3 (Cat. no. GTX110587, 1:500) were purchased from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA.
Experimental process
The cell lines were thawed by removing the cells from the vial and heated over a water bath for 1 min at 37°C. The cells were centrifuged for 4 min at 1,000 × g, and the supernatant was subsequently removed and supplemented with 10% fetal bovine serum in DMEM culture medium. The cells were cultured at a constant temperature of 37°C in an incubator with saturated humidity and 5% CO2. The cells were grown as monolayers and passaged. The medium was replaced with DMEM (10% FBS), the cells were washed with PBS for 2 times, and 0.25% pancreatin consisting of EDTA was added. The cells were then observed under an inverted microscope (Olympus Corp.), micropippetted and agitated until the cells were completely removed from the flask. When the surface of the flask was transparent and without frizz, it indicated that cells were completely detached from the flask wall. The supernatant was removed by centrifugation at 8,000 × g for 5 min and the cells were again cultured and passaged as described above. miR-184 mimics and their inhibitor were transfected into glioma cells. The LN18 cells were collected in the logarithmic phase, and the concentration of cell suspension was adjusted to 3×106/ml. The cells were cultured with DMEM (10% FBS) in 6-well plates at 37°C containing 5% CO2 for 12 h until the cell density fused to 70–80%. Then, 5 µl Lipofectamine® 2000 was added together with 200 µl serum-free culture medium and the cells were kept at a room temperature for incubation for 15 min.
Observation index qPCR detection method
Pancreatin (Boster, Wuhan, China) was used to digest the glioma cells in the logarithmic phase. The number of cells counted was approximately 5×106. Phosphate-buffered saline (PBS) was used to wash the cells, followed by centrifugation at 1,500 × g for 5 min. TRIzol reagent (1 ml) was subsequently added, and the cells were micropippetted and agitated prior to cell suspension. Chloroform (0.2 ml) was added and mixed, and the cells were again suspended prior to centrifugation. The cells were then carefully transferred to the upper aqueous phase into a new RNAase-free centrifuge tube. Without disturbing the white muddy layer, the same volume of isopropanol was added, and the mixture was suspended. The cells were again centrifuged, the supernatant was removed, and the white or transparent precipitation yielded was RNA. Subsequently, the RNA was washed in 1 ml of 75% ethyl alcohol and precipitated. The cells were again centrifuged, and the supernatant was removed. The procedure was repeated, after which RNA pellet was air-dried and dissolved in DEPC (Boster). Pre-hybridization was performed according to the instructions provided with the kit (Boster) prior to the addition of the miR-184 probe to produce the reaction. The probe was preheated with 0.2X SSC buffer (Amresco, Solon, OH, USA), rinsed, and blocked using non-specific staining. Biotinylated mouse anti-digoxin polyclonal antibody (1:200) was added and incubated at room temperature. The cells were washed with PBS and coloration was developed using DAB, with the degree of color development occurring under a microscope. The cells were then washed with tap water to terminate reaction.
MTT cell proliferation assay
Glioma cells in the logarithmic phase were collected, and seeded at a density of 5×103 cells in a 96-well plate with RPMI-1640 culture solution supplemented with 10% fetal calf serum. After culturing for 24 h the supernatant was removed. Subsequently, 20 µl sterile MTT (5 mg/ml; Sigma, St. Louis, MO, USA) was added every 24 h into the well to be measured. Three ventral orifices were set at each time point and monitored for 6 days consecutively prior to culturing for 4 h in a cell incubator. The supernatant was completely removed, and DMS0 at 150 µl/hole was added and mixed for 10 min on a shaker (Bio-Rad, Hercules, CA, USA) until the purple crystal was completely dissolved. Using a microplate reader (Bio-Rad), absorbance was measured at a wavelength of 579 nm. The IC50 of the endophytic fungi extract in in the two types kinds of self healing was measured and the experiment was repeated three times. The cell growth inhibiting rate was calculated as: (1-average value in experimental group/average value in control group) × 100%.
Clone formation
Glioma cells were collected in the logarithmic phase using regular 0.25% trypsin digestion and passaging for cell suspension. Cell suspension was diluted to a gradient, and the cells were inoculated in the culture dish with a certain density. This was followed by micropippetting and agitation, as well as dilution and inoculation. The cells were cultured for 2–3 weeks in a 37°C environment with 5% CO2, and saturated. The solution was replaced with fresh culture solution according to the change of pH. Termination of the culture occurred following identification of clone formation via visual observance. The culture solution was removed, and the cells were washed twice with PBS solution and air-dried. This process was followed by fixation with methyl alcohol for 15 min, after which the methyl alcohol was removed and the cells were air-dried. Cells were stained with the appropriate amount of Giemsa for 10–30 min, after which the staining was washed with water prior to air drying. The plate was inverted and a grid overlay of a piece of transparent film was prepared. Cell clones >50 were counted and the colony forming efficiency was calculated as: (%) = (colony number/inoculation cell number) × 100. The experiment was repeated three times.
Cell scratch and invasion assays
A line was drawn on the back of pour plate using a marker pen. Cells were added from the experimental group until each pour plate had ~5×105 cells. Using a sterile pipette, a spearhead cut was made along the bottom of the culture pour plate in a ‘―’ shape. The cells were cultured with DMEM culture solution comprising 5% calf serum and the cell migration in the scratch area was was observed. To coat the basilar membrane 50 mg/l artificial reconstructed basement membrane with Matrigel (dilution, 1:5) was used to coat the upper surface of 20 membranes (BD Biosciences, Franklin Lakes, NJ, USA) at the bottom of the Transwell chamber, and air-dried. For hydration 50 µl serum-free culture solution supplemented with 10 g/l bovine serum albumin in each plate was used. The cells were inoculated by adding 500 µl culture medium outside the chamber, and 100 µl tumor cells within the chamber. Serum-free culture medium served as the culture solution, followed by fixing and staining. Briefly, the cells were washed with PBS, and any cells on the upper layer of the microporous membrane removed using a cotton swab. The cells were then fixed with absolute alcohol and the solution was stained with crystal violet. The cells were visualized under a microscopic. The number of cells that transferred to the lower layer of the microporous membrane were counted under an inverted microscope. For each sample 10 visual fields were counted. The procedure was repeated at least three times and the average number of cells was obtained.
Western blotting
SDS-PAGE electrophoresis on 30 µg total protein and electrotransferred onto a nitrocellulose membrane (NC). The NC membrane was placed in Tris-buffered saline and Tween-20 (TBST) containing 50 g/l skim milk powder at room temperature. As per the pre-stained Marker, the NC membrane was cut into different strips based on molecular weight and inoculated in TBST solution containing p-JAK2 and p-STAT3 primary antibodies at 4°C overnight. The following day, the membrane was washed with TBST on a shaker and the strips were inoculated with 1:5,000 corresponding mouse anti-rabbit monoclonal secondary antibody. The membrane was placed into the electrophoresis gel imaging (Bio-Rad) and the images developed using ECT luminous solution. Gray value of the strips were subsequently calculated.
Statistical analysis
SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA) was applied to record and process the data. Measurement data were presented as means ± standard deviation. One-way variance analysis was applied to determine comparisons between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Comparison of the expression of miR-184, cell proliferation, clone and invasion ability
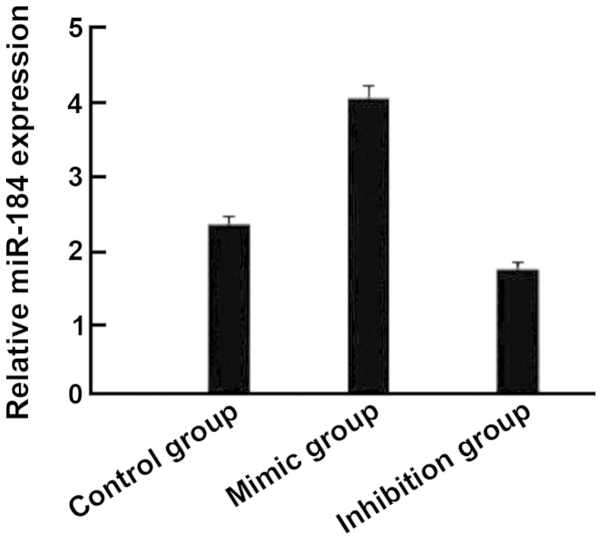
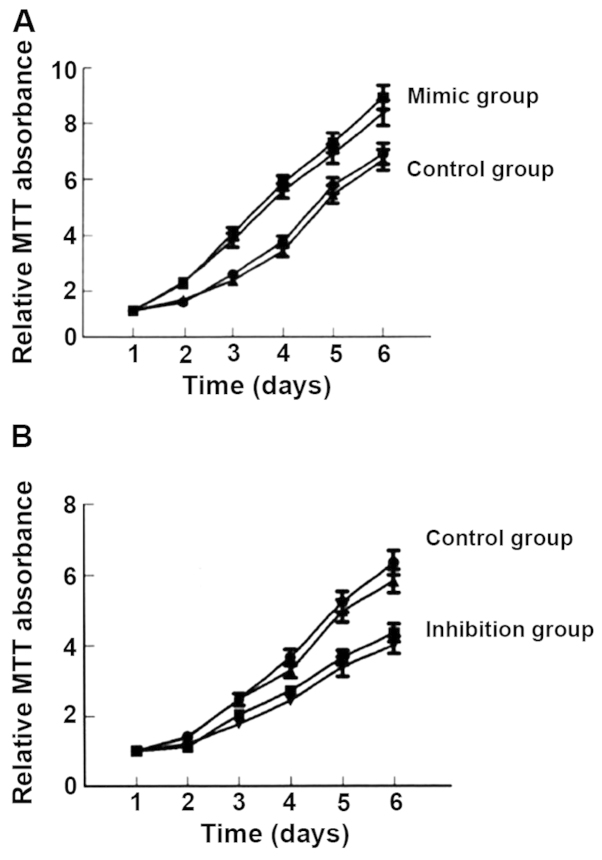
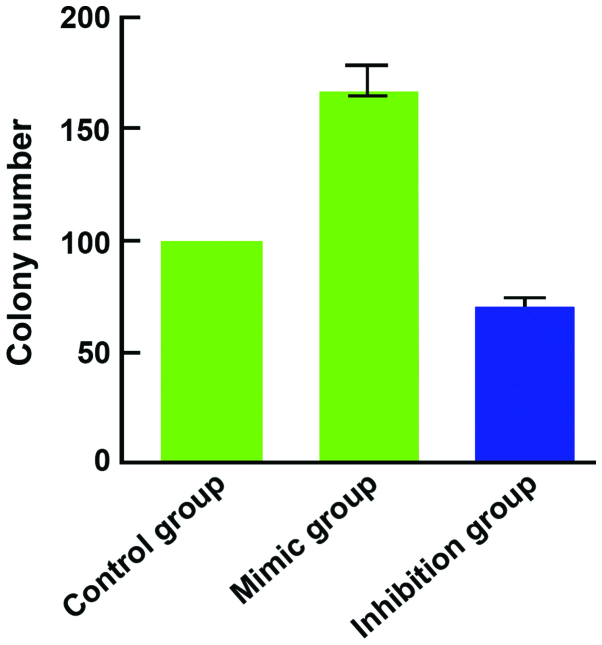
Compared with the control group, the expression of miR-184 in the miR-184 mimic group was significantly increased (Fig. 1) and cell proliferation (Fig. 2), as well as clone (Fig. 3) and invasion ability (Fig. 4) were enhanced. By contrast, the expression of miR-184 in the miR-184 inhibition group was reduced compared with the control group (Fig. 1) and cell proliferation (Fig. 2B), as well as clone (Fig. 3) and invasion ability (Fig. 4) were decreased. Differences were statistically significant (P<0.05). The number of cells permeating the septum in the control group was 98.3±12.4; in the miR-184 mimic, 115.6±15.7; and in the inhibition group, 85.7±11.3. Comparisons of the number of cells permeating the septum between groups were statistically significant (F=5.126, P<0.001) (Figs. 1–4).
Figure 1.
The relative microRNA (miR)-184 expression detected by PCR showed that the mimic group had a higher expression compared to the control and inhibition groups.
Figure 2.
Comparison of cell proliferation ability. The absorbance value for the (A) mimic group was higher than that for the control group. (B) By contrast, the absorbance value for the control group was higher than that for the inhibition group.
Figure 3.
Comparison of cell clone ability. Colony number represents the clonal ability of the cells with the mimic group, being higher that the control and inhibition groups.
Figure 4.
Comparison of cell invasion ability. The number of cell scratch test cells indicating the cell invasion ability with the mimic group exhibiting a higher number of cells compared to the control and inhibition groups.
Comparison on the expression level of p-JAK2 and p-STAT3
Compared with the control group (256.7±43.6, 243.9±34.2), the expression level of p-JAK2 (345.8±49.7) and p-STAT3 (315.2±32.1) in the mimic group was significantly higher whereas that in the inhibition group (194.2±36.9, 167.8±39.5) was significantly lower. The differences were statistically significant (P<0.05) (Table I).
Table I.
Comparison on the expression level of p-JAK2 and p-STAT3.
| Groups | p-JAK2 (%) | p-STAT3 (%) |
|---|---|---|
| Control group | 256.7±43.6 | 243.9±34.2 |
| Mimic group | 345.8±49.7 | 315.2±32.1 |
| Inhibition group | 194.2±36.9 | 167.8±39.5 |
| F | 5.627 | 6.105 |
| P-value | <0.001 | <0.001 |
JAK2, Janus kinase 2; STAT3, signal transducer and activator of transcription 3.
Discussion
MicroRNA is a type of non-protein coding micromolecule short-chain RNA that has a regulatory function and is widely identified in animals and plants. miR is closely associated with the occurrence and development of glioma (5). miR affects sequence specificity, regulates gene expression and inhibits gene expression following transcription through post-transcriptional control, thereby participating in the regulation of cell growth, proliferation, differentiation, metabolism and apotosis (6). Although a number of miR have been identifed, not all have an open reading frame. miRs may be present in eukaryotic genomes yet not encode protein. Their lengths are similar at approximately 18–22 nt. miRs belong to short sequence micromolecule RNA (7). Previous findings (8,9) have shown that the expression of several miR in glioma was different from those in normal cerebral tissue, for example, the expression of miR-21 and miR-9-2 was upregulated while that of miR-128-1, miR-181a, miR-181b, and miR-181c was downregulated. Other findings have shown that the expression of 11 types of miR was downregulated, including miR-10b and miR-21 (2). Certain miR were able to promote glioma invasion and their expression in glioma was significantly upregulated. Typical examples included miR-10-b, miR-21 and miR-221/222 (10). By contrast, certain miR were able to inhibit glioma invasion. Therefore, the role miR plays as a target for tumor diagnosis and treatment is crucial (11).
The expression of miR-184 is abnormal in various tumors, including hepatic carcinoma, lung carcinoma, nasopharyngeal carcinoma, and adrenal gland neoplasms. Additionally, miR-184 may regulate the formation mechanism of tumor through the JAK2/STAT3 signaling pathway. JAK2/STAT3 is an important signaling pathway of the JAK/STAT family. The JAK2/STAT3 channel is closely associated with inflammatory reaction, oxidative stress, cell injury and apoptosis. Lung cancer tolerance was reversed in patients undergoing radiotherapy by adopting niclosamide to destroy STAT3 phosphorylation (12). Previous findings on hematological malignancy have shown that leukemia partially resulted from overexpression or continuous activation of a number of growth factor receptors and their downstream enzymes, which were incurred from gene mutation or chromosome translocation. The joint effects of multiple channels, including JAK/STAT would lead the cells to proliferate uncontrollably and finally resulted in leukemia (13).
The results of the present study have shown that compared with the control group, the expression of miR-184 in the miR-184 mimic group was increased, and the properties of cell proliferation, clone and invasion ability were enhanced. The number of cells penetrating the septum, as well as the expression of p-JAK2 and p-STAT3 protein were increased and the differences were statistically significant. By contrast, compared with the control group, the expression of miR-184 in the miR-184 inhibition group was decreased, and cell proliferation, as well as clone and invasion ability were reduced. The number of cells penetrating the septum, as well as the expression of p-JAK2 and p-STAT3 protein were decreased and differences were statistically significant. In conclusion, miR-184 may be involved in the formation of glioblastoma and influence the expression of the JAK2/STAT3 signaling pathway.
References
- 1.Waghmare I, Roebke A, Minata M, Kango-Singh M, Nakano I. Intercellular cooperation and competition in brain cancers: Lessons from Drosophila and human Studies. Stem Cells Transl Med. 2014;3:1262–1268. doi: 10.5966/sctm.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li R, Li X, Ning S, Ye J, Han L, Kang C, Li X. Identification of a core miRNA-pathway regulatory network in glioma by therapeutically targeting miR-181d, miR-21, miR-23b, β-Catenin, CBP, and STAT3. PLoS One. 2014;9:el01903. doi: 10.1371/journal.pone.0101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Northcott PA, Fernandez-L A, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan W, Yang Y, Yan J, Yu S, Liu J, Zhou J, Zhang J, Jin Z, Yi D. The effects of curcumin post-treatment against myocardial ischemia and reperfusion by activation of the JAK2/STAT3 signaling pathway. Basic Res Cardiol. 2012;107:263. doi: 10.1007/s00395-012-0263-7. [DOI] [PubMed] [Google Scholar]
- 5.Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383:280–285. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 6.Xia H, Qi Y, Ng SS, Chen X, Li D, Chen S, Ge R, Jiang S, Li G, Chen Y, et al. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 2009;1269:158–165. doi: 10.1016/j.brainres.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 7.Garajová I, Le Large TY, Frampton AE, Rolfo C, Voortman J, Giovannetti E. Molecular mechanisms underlying the role of microRNAs in the chemoresistance of pancreatic cancer. BioMed Res Int. 2014;2014:678401. doi: 10.1155/2014/678401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saadatian Z, Masotti A, Saleh Nariman Fam Z, et al. Single-nucleotide polymorphisms within micrornas sequences and their 3′ utr target sites may regulate gene expression in gastrointestinal tract cancers. Iran Red Crescent Med J. 2014;16:el6659. doi: 10.5812/ircmj.16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orang AV, Barzegari A. MicroRNAs in colorectal cancer: From diagnosis to targeted therapy. Asian Pac J Cancer Prev. 2014;15:6989–6999. doi: 10.7314/APJCP.2014.15.17.6989. [DOI] [PubMed] [Google Scholar]
- 10.Pang JC, Kwok WK, Chen Z, Ng HK. Oncogenic role of microRNAs in brain tumors. Acta Neuropathol. 2009;117:599–611. doi: 10.1007/s00401-009-0525-0. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Duan W, Jin Z, Yi W, Yan J, Zhang S, Wang N, Liang Z, Li Y, Chen W, et al. JAK2/STAT3 activation by melatonin attenuates the mitochondrial oxidative damage induced by myocardial ischemia/reperfusion injury. J Pineal Res. 2013;55:275–286. doi: 10.1111/jpi.12070. [DOI] [PubMed] [Google Scholar]
- 12.You S, Li R, Park D, Xie M, Sica GL, Cao Y, Xiao ZQ, Deng X. Disruption of STAT3 by niclosamide reverses radioresistance of human lung cancer. Mol Cancer Ther. 2013;13:606–616. doi: 10.1158/1535-7163.MCT-13-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steelman LS, Abrams SL, Whelan J, Bertrand FE, Ludwig DE, Bäsecke J, Libra M, Stivala F, Milella M, Tafuri A, et al. Contributions of the Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways to leukemia. Leukemia. 2008;22:686–707. doi: 10.1038/leu.2008.26. [DOI] [PubMed] [Google Scholar]