Fig. 1.
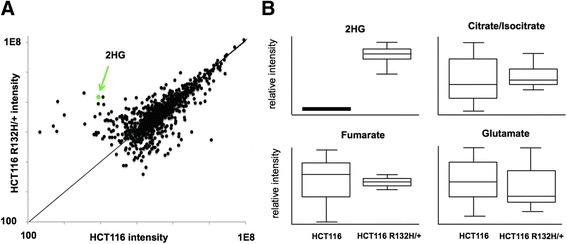
Conventional untargeted metabolomic comparison of parent HCT116 colorectal carcinoma cells to isogenic cells with heterozygous knock-in of the R132H IDH1 mutant. Results are based on n = 3 samples per group. a Each circle displayed represents a metabolomic feature detected. Features off the y = x line have altered intensities between groups. Out of the 12,481 features detected, we found that 1722 were altered with p values <0.05 and fold changes >1.5. These data support that mutant IDH1, perhaps through the production of D-2HG, affects the metabolic phenotype of colorectal carcinoma cells and therefore are a reasonable choice to study 2HG metabolism. We identified the feature shown in green as 2HG, which was increased in HCT116 cells by a fold change of 165 to approximately 3.5 mM. b Box plots comparing the levels of selected metabolites between IDH1 mutant and wild type cells
