Figure 1.
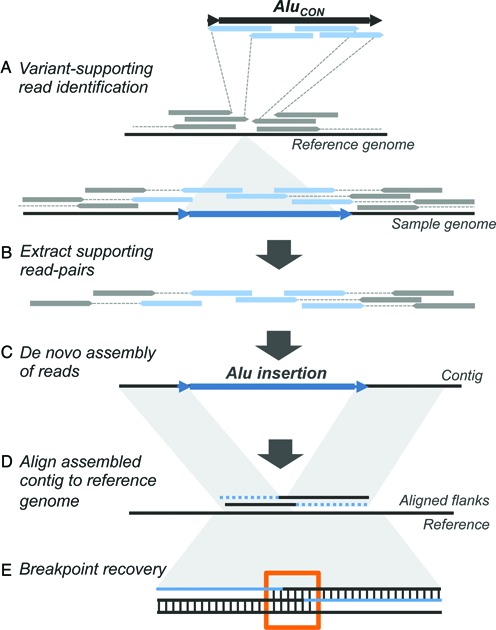
Strategy for detection and assembly of non-reference Alu insertions. Approach for reconstruction of non-reference Alu insertions from WGS data. (A) WGS in aligned BAM format from 53 samples were merged to a single BAM file, and clusters of Alu-supporting read pairs identified using the RetroSeq program by Keane et al. (B) Alu-supporting read pairs and intersecting split reads were extracted for each candidate site, and (C) Subjected to a de novo assembly using the CAP3 overlap-layout assembler (D) Alu-containing contigs were then mapped to the reference genome to verify chromosomal coordinates and uniqueness of the call. (E) Breakpoints and putative TSDs from each contig were computationally predicted by 3-way alignment to determine overlap of the assembled upstream and downstream flanks with the pre-insertion site from the hg19 reference.
