Figure 4.
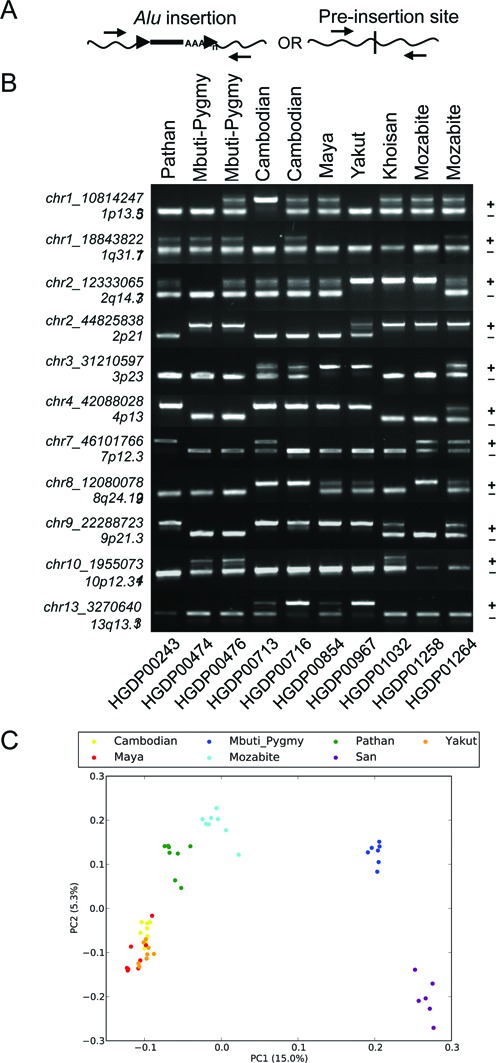
Genotyping of a subset of non-reference Alu insertions. Genotype validation was performed for 11 sites across 10 individuals. (A) Strategy for primer design and allele detection. A single primer set was used for genotyping each locus, designed to target within 250 bp of the assembled insertion coordinates relative to hg19. (B) Genotyping from PCR screens and band scoring. Banding patterns supporting the unoccupied or Alu-containing allele were assessed following locus-specific PCR; predicted band sizes were estimated by in silico PCR analysis and mapped Alu coordinates per site. The chromosomal location of each Alu is indicated at left. A ‘+’ or ‘−’ shows the relative position of each allele. Sample information is provided for population (above) and for each individual (below). (C) Principle Component Analysis (PCA) was performed on genotype matrix for 1010 autosomal sites genotypes across 53 populations. A projection of the samples onto the first two Principal Components is shown.
