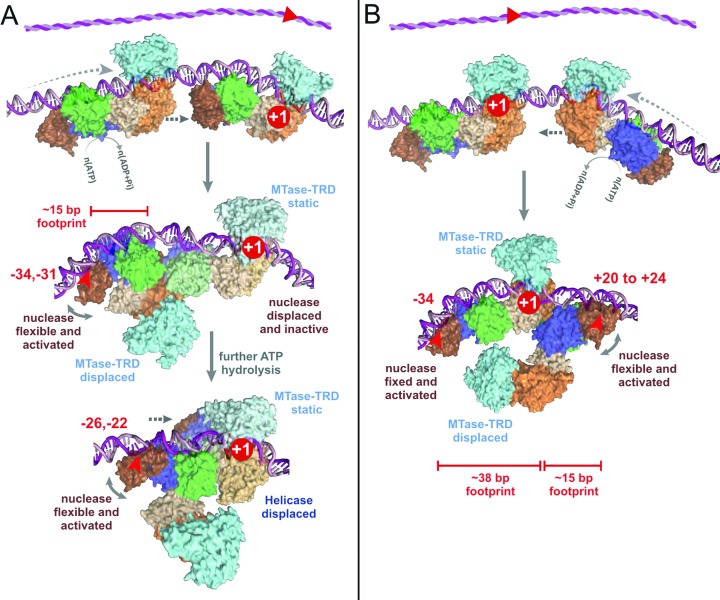
Figure 9.
Models for DNA cleavage at a site following: (A) Rear-end collision; or (B) Head-on collision. Domains coloured as in Figure 1B,1C. The red +1 represents the methylated adenine at the site and marks the static enzyme in each complex. Here, we present the MTase-TRD displacement as occurring upon collision rather than during initiation of translocation. For rear-end collision (A), the static site-bound enzyme is not activated but provides a stepwise series of roadblocks as domains peel off the DNA (Figures 4 and 7). At each roadblock, the nuclease of the rearward translocating enzyme becomes activated by motor activity, sensed through its helicase domain (Figures 3 and 4). Once the static MTase-TRD releases its site, strain is released and further nuclease activity is curtailed. Thus the nicking events localise only at the site (Figure 3). For the head-on collision of two translocating enzymes (Figure 2B and C), motor-dependent activation of the nucleases is most likely via the helicase domains in both cases. However, for head-on collision between a motile enzyme and a static enzyme at a site (B), the interacting domains are different in each enzyme: the MTase-TRD of the static enzyme which is stably associated with the DNA; and the helicase domain of the translocated enzyme because of the displacement of the MTase-TRD. Nonetheless, both nucleases become activated (Figure 5).