Figure 2.
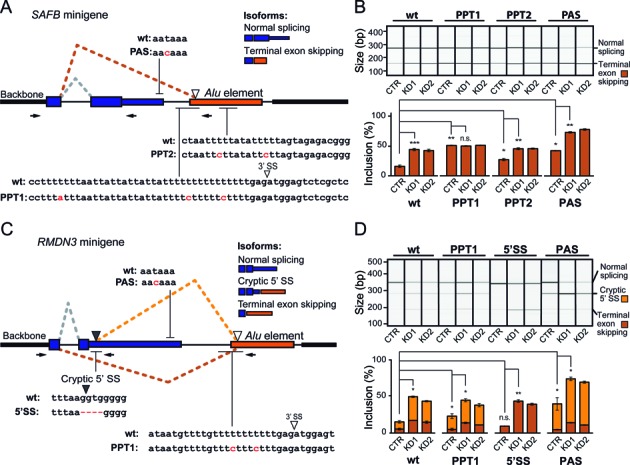
Intergenic Alu exonisation is in kinetic competition with preceding splicing and polyadenylation. (A) Schematic representation of the SAFB minigene. Under normal conditions, intergenic Alu exonisation is repressed, the terminal exon is spliced and the transcript is polyadenylated at the genuine polyadenylation site (grey dashed lines). In the HNRNPC knockdown, the intergenic Alu exon is spliced in favour of the terminal exon (dark orange dashed lines). The white arrowhead marks the 3′ splice site (3′ SS), which is recognized inside the Alu element. Depicted above and below are the nucleotide sequences in selected regions of the wild-type construct as well as the introduced mutations (red): polyadenylation signal (PAS), and upstream and downstream polypyrimidine tracts (PPT1 and PPT2, respectively). (B) Semiquantitative RT-PCR monitoring the inclusion of the intergenic Alu exon in the minigenes with wild-type (wt) or mutated sequences (PPT1, PPT2 and PAS) in HNRNPC knockdown (KD1 and KD2) and control HeLa cells (CTR). Top: Gel-like representation of the capillary electrophoresis data showing Alu exon inclusion, with the resulting isoforms indicated on the right. Bottom: Quantification of the average Alu exon inclusion. Lines indicate relevant comparisons with asterisks representing different levels of significance (*P value < 0.05; **P < 10−3; ***P < 10−4; n.s., not significant; Student's t-test). Error bars represent standard deviation of the mean, n = 3. (C) Schematic representation of the RMDN3 minigene, indicating the splicing pattern under normal conditions (grey) or the two possible scenarios of intergenic Alu exonisation upon HNRNPC knockdown (cryptic 5′ splice site activation, light orange; terminal exon skipping, dark orange). The black and white arrowheads represent the cryptic 5′ splice site in the terminal exon and the 3′ splice site within the Alu element, respectively. Nucleotide sequences and mutations as in (A): PAS, upstream polypyrimidine tract (PPT1) and cryptic 5′ splice site (5′SS). (D) Semiquantitative RT-PCR monitoring the inclusion of the intergenic Alu exon in the RMDN3 minigene with wild-type (wt) or mutated sequences (PAS, PPT1 and 5′SS) in HNRNPC knockdown (KD1 and KD2) and control HeLa cells (CTR). Gel-like representation of capillary electrophoresis (top) and quantification of average Alu exon inclusion (bottom) are shown as in (B). The two Alu exonisation scenarios (cryptic 5′ splice site activation, light orange; terminal exon skipping, dark orange) are indicated on the right.
