Figure 3.
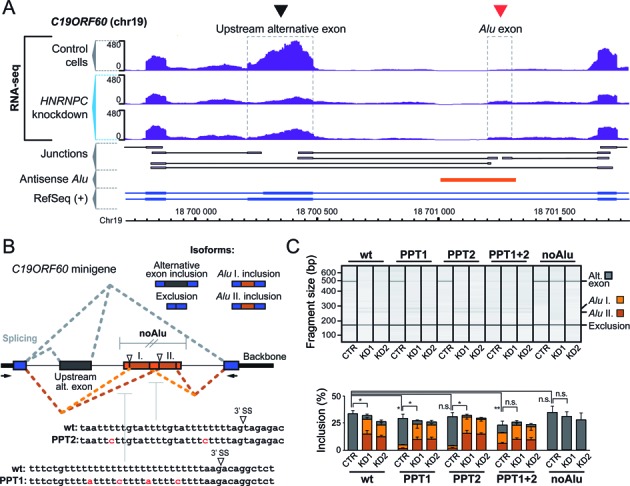
Intronic Alu exonisation interferes with the inclusion of an upstream exon. (A) Genome browser view of the C19ORF60 gene (chr19, nt 18 699 714–18 701 786, plus strand) showing the RNA-seq data from control and HNRNPC knockdown HeLa cells (labelling as in Figure 1A). The arrowheads mark the hnRNP C-repressed intronic Alu exon (red) as well as the upstream alternative exon that is downregulated upon HNRNPC knockdown (black). (B) Schematic representation of the C19ORF60 minigene, indicating the splicing pattern under normal conditions when the upstream alternative exon is either included or excluded from the transcript (grey lines), and under HNRNPC knockdown conditions when the left and right arm of the intronic Alu element exonise and the upstream alternative exon is skipped (light and dark orange lines, respectively). The white arrowheads mark the two 3′ splice sites within the Alu element. Indicated below are the nucleotide sequences and mutations for selected regions: upstream polypyrimidine tract (PPT1), downstream polypyrimidine tract (PPT2) and the combination of both (PPT1+2). The region of complete deletion of the Alu element is indicated above (noAlu). (C) Semiquantitative RT-PCR monitoring the inclusion of the intergenic Alu exon in the minigenes with wild-type (wt) or mutated sequences (PPT1, PPT, PPT1+2 and noAlu) in HNRNPC knockdown (KD1 and KD2) and control HeLa cells (CTR). Gel-like representation of capillary electrophoresis (top) and quantification of average Alu exon inclusion are shown as in Figure 2B. The different detected splicing products (inclusion of the upstream alternative exon, grey; exonisation of the first and second arm of the intronic Alu element, light and dark orange, respectively) are indicated on the right.
