Figure 6.
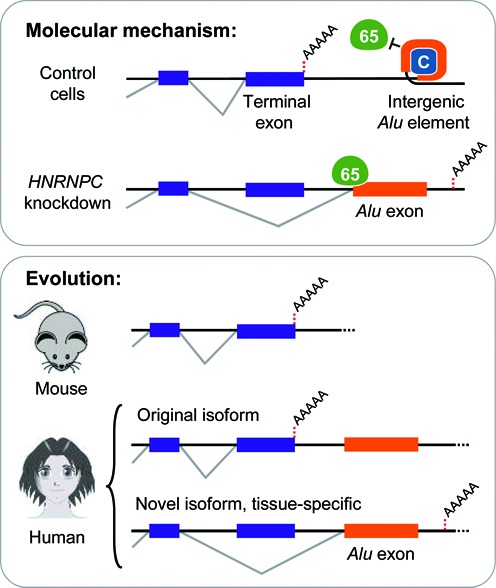
Intergenic Alu exonisation facilitated the formation of alternative transcript 3′ ends during human genome evolution. Shown is a schematic model of the hnRNP C-mediated regulation of intergenic Alu exonisation and its impact on the formation of new transcript 3′ ends during human genome evolution. Upper panel: Under normal conditions, recognition of the splice sites and exonisation of the intergenic Alu element is repressed by hnRNP C, which prevents U2AF65 from binding to the continuous U-tracts of the Alu element. Under these conditions, the terminal exon is correctly spliced and polyadenylation occurs at the genuine polyadenylation site. In the absence of hnRNP C, the splice sites of the intergenic Alu element are recognized by the splicing machinery that promotes Alu exonisation. As a consequence, the terminal exon is either skipped or spliced to the exonised intergenic Alu element through activation of a cryptic 5′ splice site inside the terminal exon (not shown). In both cases, the transcript is polyadenylated at a new downstream polyadenylation site (dashed red line). Lower panel: Schematic comparison of orthologous genes in the human genome and the Alu element-free mouse genome. In the human orthologue, transcripts either end with the conserved terminal exon (top) or contain an Alu exon downstream of last common exon (bottom).
