Figure 5.
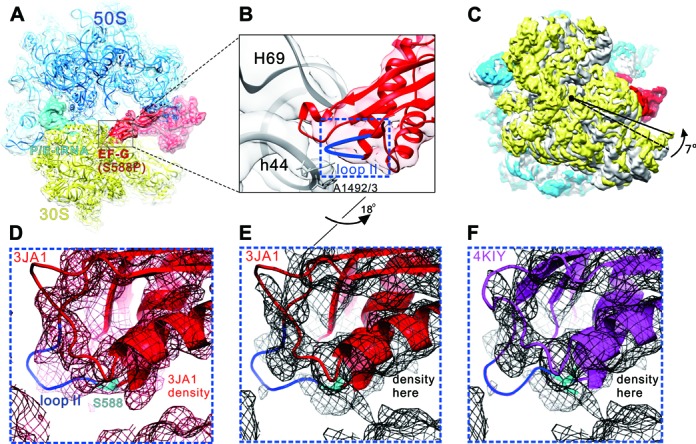
Cryo-EM structure of PoTC•EF-G (S588P)•GDPNP. (A) Overview of the PoTC with EF-G (S588P) (red) and a deacylated tRNA (cyan) in the P/E state.To model this complex, the 70S ribosome crystal structure (PDB 4KIX and 4KIY (35)) were used. (B) Zoom-in view of B2a region composed of H69 and h44 (gray), and loop II (blue) of EF-G domain IV. (C) Rotation of the 30S subunit (yellow) relative to the 50S subunit (transparent blue) caused by EF-G (S588P) binding. Compared to the classical state (gray, PDB 4V51 (50)), 30S shows a counter-clockwise rotation by 7°(viewed from the solvent side of 30S subunit). The structures are aligned using the 23S rRNA as reference. (D) Superimposition of the density map of a cryo-EM structure of 70S•EF-G (H91A) with the derived atomic model (PDB 3JA1 (41)). (E) Superimposition of our density map of the PoTC•EF-G (S588P)•GDPNP complex with the atomic model from 3JA1. (F) Superimposition of our density map of the PoTC•EF-G (S588P)• GDPNP complex with the atomic model from 4KIY (35). Compared to (B), the orientation in (D–F) has been turned ∼18° along the long axis of EF-G domain IV.
