Fig 1. Binding of LHY and CCA1 to the promoters of clock-associated genes.
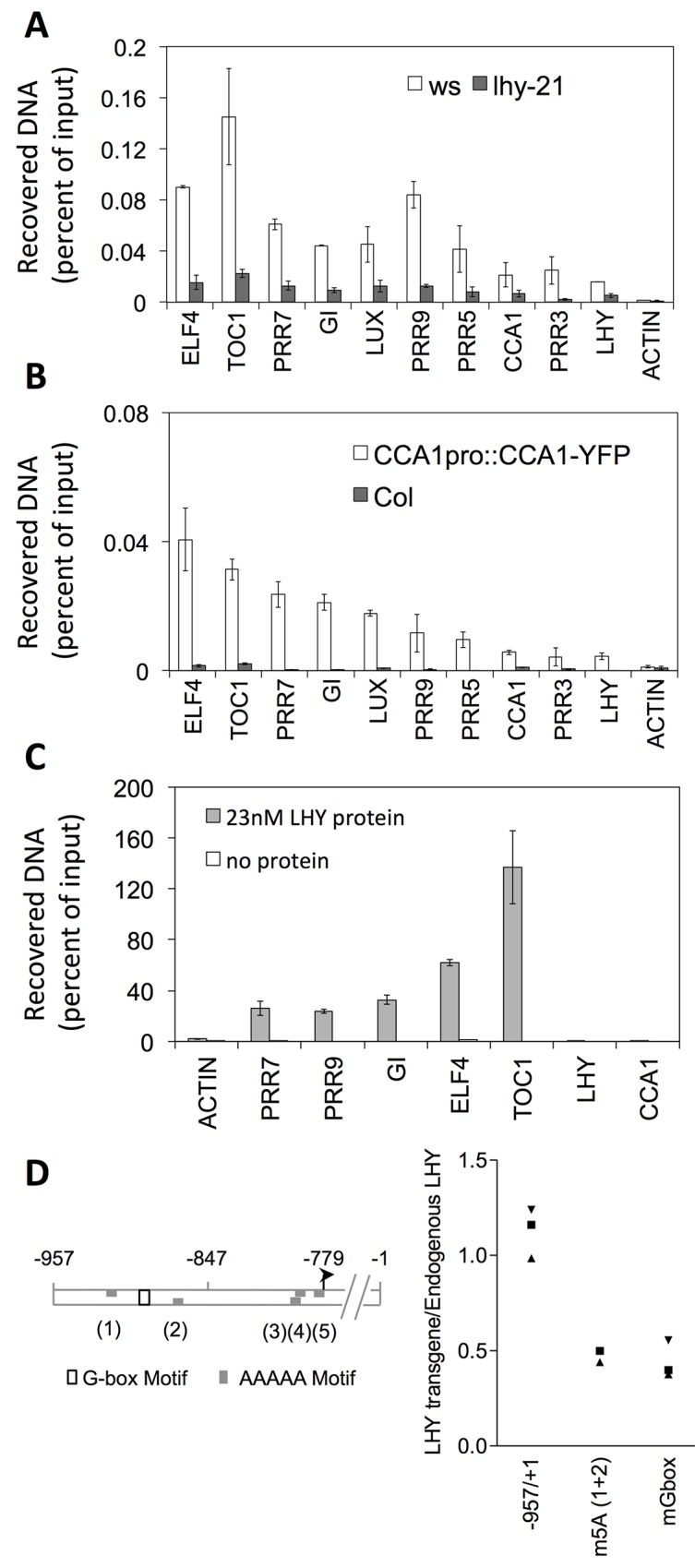
(A) In vivo binding of LHY to the promoters of clock-associated genes was tested by ChIP-qPCR analyses of wild-type samples using a polyclonal antibody to the LHY protein. Plants were grown under 12L12D light-dark cycles and tissue was sampled two hours after dawn, corresponding to the time when LHY protein levels are at their maximum [31]. The ACTIN locus was used as a negative control. In order to demonstrate the specificity of the antibody, ChIP enrichments for wild-type (Ws) plants were compared to those for the insertion mutant, lhy-21. Error bars represent standard errors from two independent biological experiments. (B) In vivo binding of CCA1 was tested by performing ChIP-qPCR analyses on cca1-1 CCA1pro::CCA1-HA-YFP or wild-type plants using an antibody to the YFP tag. Enrichment for the ACTIN2-7 locus is shown as a negative control. Error bars represent standard errors from two independent biological experiments. (C) In vitro binding of LHY to purified genomic DNA. Bacterially expressed, His-tagged LHY protein was used to pull down sheared, purified genomic DNA. The resulting enrichment for different target promoter sequences was quantified by real time PCR using the same primers as used for ChIP analyses, and expressed as a percentage of input material. This experiment detected binding to all LHY binding targets identified in ChIP experiments, except for the LHY and CCA1 promoters. Error bars represent standard deviations from two independent binding experiments. (D) Mutations of the G-box and 5A sites disrupt in vivo binding of the LHY protein to its own promoter. The diagram at the top of the panel shows the relative positions of G-box and 5A motifs in the LHY promoter. Positions are numbered relative to the ATG and the arrow represents the transcriptional start site. ChIP experiments were carried out to assay binding of the LHY protein to wild-type and mutated LHY::luc transgenes. In order to account for differences in sample preparation, enrichment for LHY::luc transgene sequence was expressed relative to endogenous LHY promoter sequence. Relative enrichment levels close to 1 were obtained using the wild-type construct (-957/+1), indicating equivalent enrichment for the endogenous and the transgenic copies of the LHY promoter. However, mutation of the G-box flanking regions (ACCACGTGTC to GTCACGTGAC) reduced binding of the LHY protein by over 50%. Mutation of both flanking 5A sites [5A (1) CCAAAAA to TGTCAAA and 5A(2) TTTTTCC to TTTGACA] had a similar effect. Each data point represents the relative enrichment for one transgenic line. Error bars from Q-PCR analyses have been omitted for clarity.
