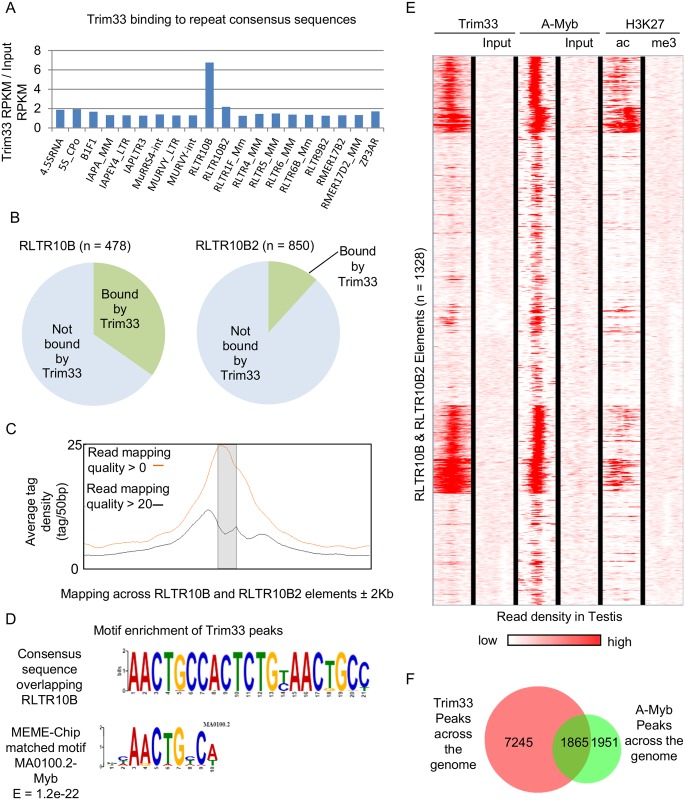
Fig 3. Trim33 binding at RLTR10B and RLTR10B2 elements.
(A) Trim33 enrichment at retrotransposon classes that show at least a 1.25 fold change over input. RLTR10B and, to a lesser extent RLTR10B2, are enriched in Trim33 binding. (B) Trim33 binds to subsets of RLTR10B (166/478) and RLTR10B2 (100/850) repeats in the genome. (C) Read density for Trim33 is shown across the RLTR10B and RLTR10B2 element unit (shaded) ± 2Kb. All reads are shown in red and separately all reads with a Bowtie2 read quality of > 20 are shown in black. (D) The MEME-chip tool was used to identify enriched motifs in the sequences that overlapped Trim33 binding peaks across the genome in testis. The most significant of these matched a Myb transcription factor consensus sequence within RLTR10B elements. (E) Heat plot of ChIP-seq read density, clustered by similarity, for Trim33 and input for all RLTR10B and RLTR10B2 elements, against publically available ChIP-seq data for the A-Myb (plus input), GEO accession GSE44690, in testis and the testis Encode datasets for H3K27ac and H3K27me3. Each element shown is generated from 1Kb either side of the centre of RLTR10Bs and RLTR10B2s in the genome. (F) Significant Trim33 ChIP-seq peaks across the genome were compared to significant A-Myb ChIP-seq peaks [18] across the genome, the overlap is shown.