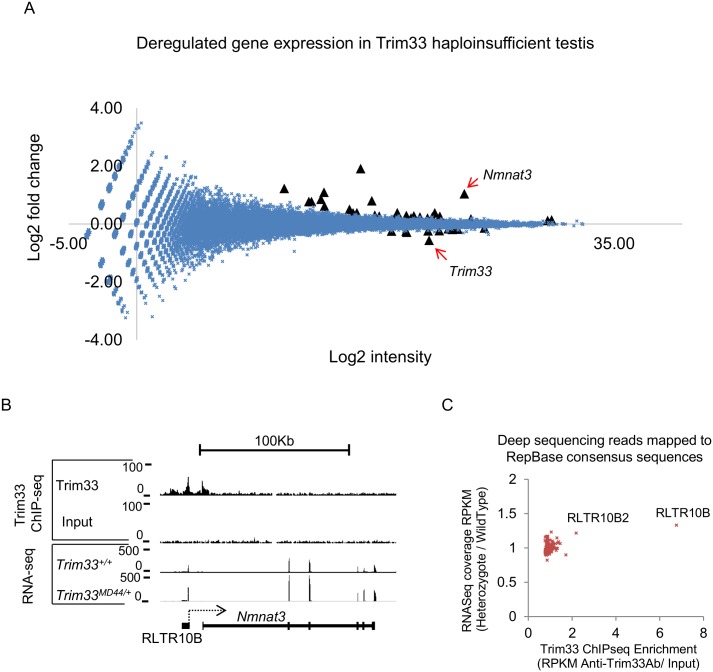
Fig 5. Trim33 regulates a group of genes in the testis.
(A) The Mean Intensity plot of RNA-seq data shows the log2 fold change of transcripts between wildtype (n = 3) and heterozygotes (n = 4), along a log2 scale of expression intensity (i.e. wildtype levels multiplied by heterozygous levels). Black triangles represent the 39 genes predicted to be significantly differentially expressed between the two sample groups. Trim33 is indicated as significantly lower in heterozygotes. (B) Trim33 binding at Nmnat3. Visual inspection of the RNA-seq reads suggested that this gene was expressed from an upstream RLTR10B. (C) Reads from each individual repeat were mapped to a Repbase consensus sequence. Those repeat classes at which the RPKM was < 10 were excluded. The ratio of RPKM in wildtypes versus that in heterozygotes is shown on the Y-axis. Trim33 ChIP enrichment at each repeat class is shown on the X-axis. Only one class, RLTR10B (indicated), was bound by Trim33 (seven fold enriched) more than threefold above Input values and was at least 1.3 fold upregulated in heterozygous mutant mouse testis.