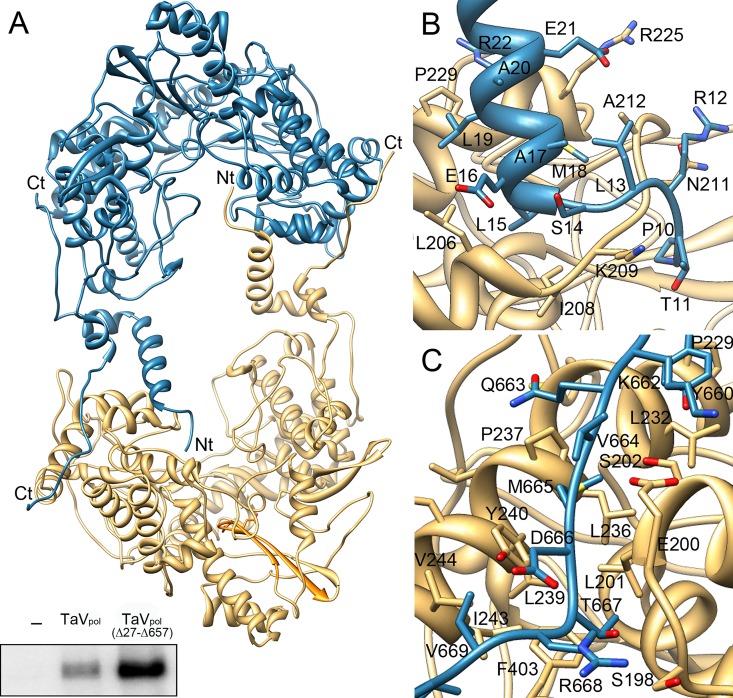
Fig 3. Structure of the TaVpol dimer.
(A) The two molecules of the crystal asymmetric unit are shown as ribbons in blue and wheat, respectively. Amino acids at the N- and C-terminal ends maintain the dimeric structure of the enzyme. The bottom inset shows in vitro polymerization assays evidencing that deletion of the first 27 N-terminal amino acid residues enhances enzymatic activity. (B) Close-up of interactions established between the N-terminus of the molecule B (blue) with the central cavity of molecule A (wheat). The first nine protein residues are disordered and not visible in the electron density. The first visible residue, P10, is located at a 18 Å of the active site. (C) Close-up of interactions established between the C-terminus of the molecule B with the fingers domain of molecule A. In panels B and C, the side chains of residues involved in intermolecular interactions are depicted as sticks and explicitly labeled. Due to the presence of the non-crystallographic symmetry (ncs) dyad interactions between the N- and C-terminus of molecule A with molecule B and those of the N- and C- terminus of molecule B with molecule A are identical.