Extended Data Figure 1. Affinity purification of spliceosomes with Prp19 requires reconstitution with U6 snRNA and enhances the potential to detect rescue by thiophilic metals.
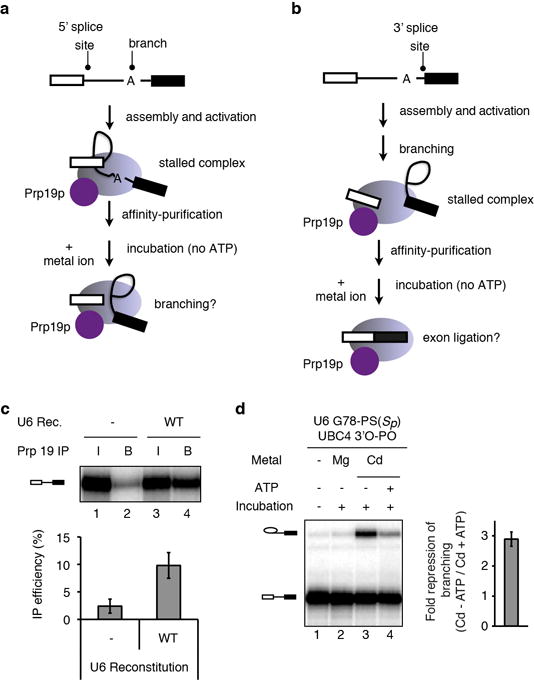
a–b, Schemes depicting experimental strategy for staging spliceosomes to monitor branching (a) or exon ligation (b) in the absence of ATP. Spliceosomes are depicted as light magenta ovals and Prp19p as a magenta circle. Following affinity purification of spliceosomes via Prp19p (ref. 36), beads were washed to remove ATP and soluble factors, and metals ions were added to assay for splicing. c, Prp19p-mediated affinity purification of activated spliceosomes, reflected by immunoprecipitation of pre-mRNA, is specific for properly reconstituted complexes. Note that the affinity purification allows quantification of the branching efficiency for activated complexes, independently of any effects on assembly. RNA from 10% of the reaction (input, I) or from beads after affinity purification (B) was extracted and analyzed by denaturing PAGE. (top) raw data; (bottom) quantification of IP efficiency; Rec., reconstitution. d, ATP represses the Cd2+ rescue for G78-PS(Sp) spliceosomes. Spliceosomes were assayed as in Fig. 2e; for lane 4, 2mM ATP-Mg2+ was also present during the incubation. (left) representative gel; (right) quantification of the extent of ATP repression. Values are averages; error bars, s.d. (n=3).
