Extended Data Figure 3. A divalent metal binds the 5′ splice site pro-Rp oxygen during branching.
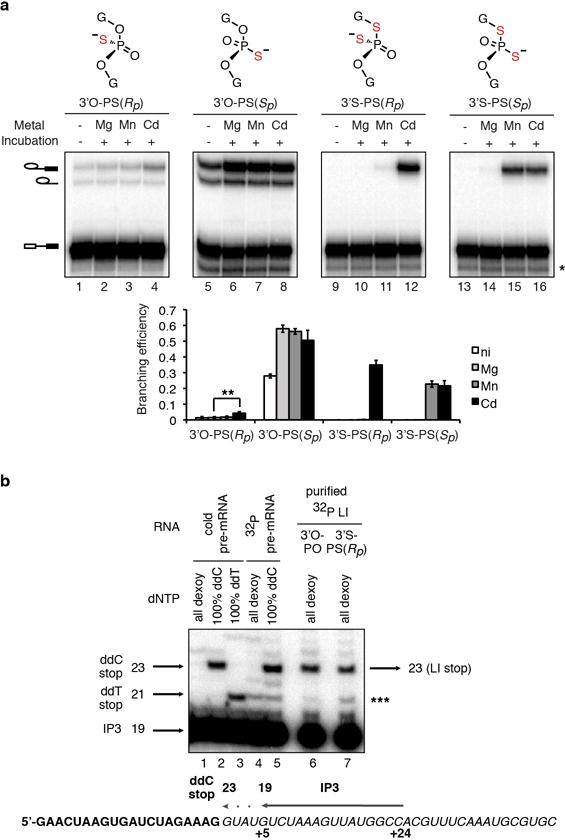
a, UBC4 pre-mRNAs bearing the indicated modifications at the 5′ splice site (top panel) were assayed as in Fig. 3a. The band marked * results from 5′ to 3′ exonucleolytic degradation that is blocked by the sulfur; ** denotes statistical significance of Cd2+ rescue compared to Mg2+ splicing (p = 0.004, paired, 1-tailed t-test, n=3). The data from Fig. 3a are reproduced here to aid comparison. Values are averages; error bars, s.d. (n=3). b, Mapping of the site of branching. Purified, 32P-labeled, intact lariat intermediates (LI ~ 0.2 fmol) resulting from branching of the UBC4 3′O-PO or 3′S-PS(Rp) substrates were used as templates for reverse transcriptase using primer IP3 (~10 fmol), which binds at nucleotides +5 to +24 of the intron (see lower diagram). Lanes 6 and 7 show that the major RT stop occurs at the same position when either the 3′O-PO or 3′S-PS(Rp) lariats are used as template. This stop migrates at the expected position, which is the position of the ddC stop resulting from extension of primer IP3 with pre-mRNA as template and at the expected position and therefore corresponds to position +1 of the intron, the expected branch site. Lower diagram shows mapping of the primer and expected RT stop onto the UBC4 pre-mRNA sequence; bold exon; italics intron. Note that the band marked *** is present in the 32P pre-mRNA lane and thus is likely a nonspecific band resulting from contamination with pre-mRNA degradation products that can anneal to the primer and serve as template.
