Abstract
Cancer-predisposing genes associated with inherited cancer syndromes help explain mechanisms of sporadic carcinogenesis and often inform normal development. Cowden syndrome (CS) is an autosomal-dominant disorder characterized by high lifetime risks of epithelial cancers, such that ∼50% of affected individuals are wild-type for known cancer-predisposing genes. Using whole-exome and Sanger sequencing of a multi-generation CS family affected by thyroid and other cancers, we identified a pathogenic missense heterozygous SEC23B variant (c.1781T>G [p.Val594Gly]) that segregates with the phenotype. We also found germline heterozygous SEC23B variants in 3/96 (3%) unrelated mutation-negative CS probands with thyroid cancer and in The Cancer Genome Atlas (TCGA), representing apparently sporadic cancers. We note that the TCGA thyroid cancer dataset is enriched with unique germline deleterious SEC23B variants associated with a significantly younger age of onset. SEC23B encodes Sec23 homolog B (S. cerevisiae), a component of coat protein complex II (COPII), which transports proteins from the endoplasmic reticulum (ER) to the Golgi apparatus. Interestingly, germline homozygous or compound-heterozygous SEC23B mutations cause an unrelated disorder, congenital dyserythropoietic anemia type II, and SEC23B-deficient mice suffer from secretory organ degeneration due to ER-stress-associated apoptosis. By characterizing the p.Val594Gly variant in a normal thyroid cell line, we show that it is a functional alteration that results in ER-stress-mediated cell-colony formation and survival, growth, and invasion, which reflect aspects of a cancer phenotype. Our findings suggest a different role for SEC23B, whereby germline heterozygous variants associate with cancer predisposition potentially mediated by ER stress “addiction.”
Introduction
Cowden syndrome (CS [MIM: 158350]) is an underdiagnosed difficult-to-recognize autosomal-dominant disorder characterized by multiple hamartomas and an increased predisposition to breast, thyroid, endometrial, and other cancers. Other, non-neoplastic phenotypes include macrocephaly and mucocutaneous findings.1, 2 The tumor-suppressor gene phosphatase and tensin homolog (PTEN [MIM: 601728]) was identified as the first susceptibility gene for CS.3, 4 Earlier studies identified germline PTEN mutations in 85% of CS individuals1, 5 meeting the strict International Cowden Consortium (ICC) operational diagnostic criteria, which select for familial cases and exaggerated phenotypes accrued from tertiary medical centers.6, 7 Subsequent prospective community-based accrual of >3,000 probands over 12 years revealed that ∼25% of individuals who met the diagnostic criteria harbored germline pathogenic PTEN mutations.8 Relatedly, we note that many individuals show clinical features reminiscent of CS but do not meet the ICC criteria. We refer to this phenotype as CS-like (CSL), and these affected individuals show extensive phenotypic heterogeneity, and only 5% have germline PTEN mutations.1
Relevant to clinical practice, we generated a predictive PTEN risk calculator8 that evaluates the pretest probability that an individual harbors a deleterious germline PTEN mutation on the basis of key clinical features.8 The resultant score is known as the PTEN Cleveland Clinic score, or CC score. A CC score of 10, corresponding to a pretest probability of >3% and a sensitivity of 90%, is recommended as a threshold for referring adult CS or CSL individuals for PTEN mutational testing and further genetic assessment. However, many CS and CSL individuals evaluated in clinic have high CC scores (range 10–54) and no underlying germline PTEN mutations or large deletions, indicating high phenotypic burden independent of PTEN and, hence, a high likelihood that other susceptibility genes exist. Indeed, we have so far reported that within this PTEN-mutation-negative subset of CS and CSL individuals, up to 8% harbor germline variants in mitochondrial complex II subunit genes SDHB (MIM: 185470), SDHC (MIM: 602413), and SDHD (MIM: 602690),9, 10 up to 30% have promoter hypermethylation in tumor-suppressor gene KLLN (MIM: 612105),11 and 11% have germline AKT1 (MIM: 164730) or PIK3CA (MIM: 171834) gain-of-function mutations.12
Epithelial thyroid carcinomas (both papillary and follicular histologies) are a major component of CS.13 We have shown in a prospective series of 2,723 CS and CSL individuals that germline mutations in a subset of the established susceptibility genes (PTEN, SDHB, SDHC, SDHD, and KLLN) increase the risk of epithelial thyroid cancer by at least 45-fold in comparison to the risk in the US general population.14 Moreover, included in our series of CS individuals are PTEN-mutation-positive probands diagnosed under the age of 18 years (the earliest age at diagnosis was 7 years), reflecting a strong genetic component in the etiology of thyroid cancer. In support of these findings, epidemiological studies have reported that thyroid cancers show a high index of “familiality” among all solid tumors.15, 16 However, the hunt for candidate genes associated with familial thyroid cancer continues today, and there is an increased appreciation for the heterogeneity of the disease and the contribution of hereditary predisposition and the micro- and macro-environment to tumor pathobiology.17 In this context, not only would additional CS-relevant genes serve as candidates for apparently sporadic predisposition to epithelial thyroid cancer, but their identification would also improve molecular diagnosis, especially predictive testing, cancer risk assessment, genetic counseling, and clinical management in CS and CSL. Therefore, we sought to identify previously undescribed cancer-predisposing gene(s) in CS and CSL via an approach combining exome sequencing and family studies and, subsequently, to characterize the functional consequence.
Material and Methods
Research Participants and Clinical Data
We scanned our clinical database for accrued CS- and CSL-affected families. Eligible families consisted of at least one unaffected and two affected family members. Eligible probands tested negative for germline mutations in PTEN, SDHB, SDHC, SDHD, AKT1, and PIK3CA and met at least the relaxed ICC operational diagnostic criteria (Table S1). Relaxed criteria are defined as full criteria minus one, and such individuals are referred to as having CSL.7
For all selected individuals, we reviewed the CC score, a semiquantitative score that is based on weighting clinical features and that estimates the pretest probability of finding a germline PTEN mutation (see Web Resources).8 Given that all individuals are wild-type for PTEN, we used the CC score as a surrogate of phenotypic burden. We hence targeted probands with a high CC score and eligible family members with available blood samples for genetic testing. We reviewed medical records, including pathology reports, for each research participant and extracted family history from clinical notes associated with cancer genetics and/or genetic-counseling visits, where applicable and with the individuals’ consent. Cleveland Clinic Institutional Review Board approval (protocol 8458) and informed consent from all research participants were obtained for this study.
Exome Sequencing and Bioinformatic Analysis
We subjected germline genomic DNA extracted from peripheral-blood leukocytes of the eligible proband to exome sequencing. Exome enrichment was performed with the TruSeq SBS v.3 Kit (Illumina), and subsequent 100 bp paired-end sequencing was performed with an Illumina HiSeq 2000 platform. Sequencing was performed at an Illumina Sequencing Service Center. Raw sequencing reads were mapped to the human reference haploid genome sequence (UCSC Genome Browser hg19) with the Burrows-Wheeler Aligner (BWA v.0.6.1).18 Insertion or deletion (indel) realignment, base- and quality-score recalibrations, and removal of PCR duplicates from the resultant binary alignment map (BAM) files were performed with the Genome Analysis Toolkit (GATK),19, 20 Sequence Alignment/Map (SAMtools), and Picard.21 Variant discovery and genotype calling of single-nucleotide variations (SNVs) and short (<50 bp) indels were performed with the GATK Haplotype Caller.
Variant Filtration and Annotation
To prioritize causal variants, we applied the ANNOVAR Variants Reduction pipeline.22 First, we discarded synonymous variants and intronic variants more than 2 bp from exon boundaries. We retained variants in conserved genomic regions on the basis of a 46-species alignment and removed variants existing in segmental duplication regions. We excluded variants with a minor allele frequency (MAF) ≥ 0.0005 in the 1000 Genomes Project (April 2012 release) or the NHLBI Exome Sequencing Project (ESP) Exome Variant Server (ESP6500 dataset) and variants reported in the dbSNP137 non-flagged database (excluding clinically associated SNPs).
To predict the potential impact of missense variants on protein function, we used the program Consensus Deleteriousness (Condel v.1.5),23 which combines multiple prediction software algorithms (SIFT,24 PolyPhen-2,25 and MutationAssessor26) into a single weighted score. We inspected all resultant variants through the Integrative Genomics Viewer (IGV).27, 28 We also used an in-house database containing variants from 11 individuals with sporadic polyposis and 13 exomes from individuals with unrelated phenotypes (connective-tissue disorders); these were sequenced with our cohort of research participants as an additional internal control filter. We excluded variants that appeared more than once in these in-house exome databases.
Mutation Validation
Mutations in candidate genes of interest were validated by PCR-based region-specific mutation analysis through Sanger sequencing. In brief, gene-specific primers encompassing exonic regions harboring the particular variations were designed. Sequencing was performed in the forward and reverse directions with Applied Biosystems ABI 3730xl DNA Analyzers at the Genomics Core of the Lerner Research Institute of the Cleveland Clinic and the Eurofins Genomics DNA-sequencing facility (Louisville). Resultant chromatograms were analyzed with Mutation Surveyor DNA Variant Analysis Software (SoftGenetics), and mutations were reported according to the Human Genome Variation Society (HGVS) guidelines. Incidental findings were reported to our genetic counselors to be communicated to the respective affected individuals and physicians of record according to the ACMG guidelines.29 Confirmed variants were sequenced in all recruited family members regardless of disease status.
Variant Interpretation and In Silico Pathogenicity Predictions
Variants retained after prioritization were considered potentially pathogenic. Importantly, prioritized variants had to be shared in all affected members of the pedigree. Further prioritization was performed on the basis of the relation of the genes to the clinical phenotypes (where applicable), predicted loci of the mutations affecting functional protein domains, and in silico assessment of the mutations’ effect on protein integrity through a combination of mutation-prediction algorithms, namely Condel,23 SIFT,24 PolyPhen-2,25 MutationTaster,30 MutPred,31 and Combined Annotation Dependent Depletion (CADD).32
Prioritized candidate genes resulting from pedigree analysis were then screened in an independent series of 96 CS and/or CSL probands who were negative for germline mutations in PTEN, SDHB, SDHC, and SDHD and who had overlapping clinical phenotypes with those observed in the pedigree under study (mainly thyroid, breast, and endometrial cancers). Gene-specific primers were designed for all exons, and Sanger sequencing was performed as stated above.
The accession numbers for the C16orf72, PTPN2, and SEC23B sequences reported in this paper are GenBank: NM_014117.2, NM_080422.2, and NM_001172745.1, respectively.
TCGA Germline Variants Dataset Analysis
We analyzed The Cancer Genome Atlas (TCGA) datasets on papillary thyroid carcinoma (THCA, n = 494) and a subset of breast invasive carcinoma (BRCA, n = 222) and uterine corpus endometrioid carcinoma (UCEC, n = 156). We analyzed all tumors with sequenced adjacent normal tissue or blood for analysis of germline mutations. Germline mutations were extracted and analyzed from raw sequencing data. In brief, genomic loci corresponding to candidate genes were extracted from the aligned BAM files, and a MAF ≤ 0.01 was used for filtering variants through dbSNP137, 1000 Genomes, and ESP6500. Mutation predictions were obtained from SIFT,24 PolyPhen-2,25 and MutationTaster.30 Importantly, we visualized and manually curated all mutations through the IGV27, 28 to confirm that they indeed exist in the tumor and adjacent normal tissue and/or blood and are hence in the germline. Corresponding clinical data were obtained from the TCGA Data Portal Matrix.
Predicted Structural Effects of Variants
We predicted the structural effects of variants by using the HOPE server, which integrates a series of standard algorithms to generate predictions.33 Because the Homo sapiens SEC23B crystal structure has not been resolved, the crystal structure of the human SEC23A-SEC24A heterodimer, complexed with the SNARE protein SEC22B34 (PDB: 2NUP), was identified as a modeling template for predicting structural effects.
Cell Lines and Culture Conditions
Immortalized lymphoblastoid cell lines (LCLs) derived from affected individuals or normal control individuals were generated by the Genomic Medicine Biorepository of the Genomic Medicine Institute of the Cleveland Clinic according to standard procedures and were subsequently maintained in RPMI-1640 supplemented with 20% fetal bovine serum (FBS) and 1% penicillin and streptomycin. HEK293T cells (originally purchased from the American Type Culture Collection) in 2011 and obtained in 2014 from the Cleveland Clinic Lerner Research Institute Cell Culture Core) were cultured in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin. The Nthy-ori 3-1 human thyroid follicular epithelium cell line (catalog no. EC90011609, lot no. 09C008, passage no. 16, purchased in 2014 from Sigma) was cultured in RPMI-1640 supplemented with 2 mM glutamine and 10% FBS. All cell lines were maintained at 37°C and 5% CO2 culture conditions and tested negative upon routine mycoplasma testing with the MycoAlert Mycoplasma Detection Kit (Lonza) at the C.E. lab (luminescence ratios < 0.9). Cell lines used have not been listed as cells known to be misidentified according to the International Cell Line Authentication Committee.35 The Nthy-ori 3-1 cell line was authenticated through short-tandem-repeat PCR (AmpFLSTR SGM Plus PCR Amplification Kit, Life Technologies) by the European Collection of Cell Cultures (original source of the cell line, test date April 14, 2009).
RNA Extraction and qRT-PCR
RNA was extracted from LCLs derived from affected and unaffected individuals with the RNeasy Mini Kit (QIAGEN), purified with Turbo DNase treatment (Life Technologies), and reverse transcribed with Superscript III Reverse Transcriptase (Life Technologies). Primers were designed for gene transcripts of interest, and cDNA was quantified with SYBR Green (Life Technologies). Results were analyzed by the standard ΔΔCT method.
Immunoblotting
Protein was extracted from whole-cell lysates with the Mammalian Protein Extraction Reagent M-PER (Thermo Scientific Pierce) supplemented with a cocktail of protease and phosphatase inhibitors (Sigma-Aldrich) and was quantified with the BCA Protein Assay Kit (Thermo Scientific Pierce). Lysates were separated by SDS-PAGE and transferred onto nitrocellulose membranes. We probed for anti-SEC23B rabbit polyclonal (Abcam ab151258) at 1:1,000, for anti-SEC23A rabbit polyclonal generated against synthetic peptides (CQKFGEYHKDDPNSFRFSET and DNAKYVKKGTKHFEA)36 at 1:2,000, and for anti-GAPDH rabbit monoclonal (Cell Signaling 2118) at 1:20,000 dilutions. Blots were scanned digitally and quantified with the Odyssey Infrared Imaging System (Li-Cor Biosciences).
Plasmids, Mutagenesis, and Cell-Line Transfection and Transduction
GFP- and FLAG-tagged wild-type SEC23B plasmids (pMSCV and pEGFP-C3 vectors, respectively) were used.36 Wild-type plasmids were mutagenized for the missense mutation of interest with the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). All expression constructs were validated by Sanger sequencing prior to transfection or transduction. For transient overexpression, cell lines were transfected with Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s instructions. Fresh media replaced the transfection media 5 hr after transfection. Cells were grown for at least 24 hr before harvesting. To generate stable cell lines, we kept retrovirally transduced cells (a pool, given that no individual clones were isolated) under 1 μg/ml puromycin selection for >30 days prior to downstream interrogation. We used four different pools of cells at passage no. 28 after selection. All experiments were conducted with cells at passage nos. between 29 and 42.
Immunofluorescence
Cells were seeded on coverslips and were fixed with 4% paraformaldehyde for 10 min at room temperature. Coverslips were mounted with ProLong Gold Antifade mountant with DAPI (Invitrogen). Slides were visualized and images were obtained with a TCS SP5 confocal microscope (Leica).
Immunoprecipitation
Cells were pelleted after transfection and lysed with M-PER (Thermo Scientific Pierce) supplemented with a cocktail of protease and phosphatase inhibitors (Sigma-Aldrich). Protein lysates were collected by centrifugation at 13,000 RPM for 15 min at 4°C and pre-cleared by incubation with protein A/G PLUS-Agarose immunoprecipitation reagent (Santa Cruz sc-2003) for 30 min at 4°C on a rotator. Pre-cleared protein lysates were quantified with the BCA Protein Assay Kit (Thermo Scientific Pierce), and 1 mg/ml lysates were prepared. We used anti-SAR1A (Santa Cruz sc-130463) and anti-DDK (Origene 4C5) antibodies for pull-down and immunoblotting at the recommended dilutions.
Doubling Time and Cell Viability
Cells were trypsinized, homogenized, and counted with the Countess Automated Cell Counter (Invitrogen). We used Trypan blue to account for dead cells and to assess cell viability. We also used the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (Sigma) to assess cell viability according to the manufacturer’s instructions.
In Vitro Migration and Invasion Assays
Transiently transfected cells were grown for 48 hr, and a migration assay was performed according to Liang et al.,37 with the minor variation of creating scratches on the cell monolayer with a p10 pipet tip. Migration of the stable cell lines was assessed with 6.5 mm transwells and 8.0 μm pore polycarbonate membrane inserts (Corning). Invasion was assessed with BioCoat Matrigel invasion chambers and 8.0 μm polyethylene terephthalate (PET) membrane (Corning). Cells were serum starved for 24 hr and harvested with the HyClone HyQTase Cell Detachment Reagent (GE Healthcare). For both assays, 5 × 104 cells were seeded in the upper chambers in serum-starved media. The lower chambers were filled with complete growth media supplemented with 10% FBS; however, for background correction sets, both chambers were filled with serum-starved media. Migration and invasion experiments proceeded for 24 hr. Cells that migrated or invaded were fixed with 4% paraformaldehyde for 5 min, permeabilized with 100% methanol for 20 min, and stained with Giemsa for 15 min all at room temperature. Cells were photographed under 10× magnification and counted in quadruplicate fields of view in triplicate membranes.
SDS-PAGE Zymography
Cells were serum starved for 24 hr, and conditioned media were collected and concentrated with spin columns with a 10 kDa molecular-weight cutoff (Biovision). Protein concentrations were determined with the BCA Protein Assay Kit (Thermo Scientific Pierce). Equal volumes of sample and zymogram sample buffer (Bio-Rad) were mixed and loaded on 10% gelatin zymogram gels (Bio-Rad). After electrophoresis, gels were washed with renaturation buffer (Bio-Rad), incubated for 18 hr at 37°C in developing buffer (Bio-Rad), and stained for 2 hr with Coomassie Brilliant Blue R-250 staining solution (Bio-Rad). Enzymatic activity was visualized as a colorless band against the Coomassie blue background. Bands were quantified with Image J software (NIH), and values were normalized to total protein.
ER-Stress Induction and Assessment of Cell Viability and Colony Formation
Cells were seeded and allowed to grow overnight and then treated with either 1 μM thapsigargin or 0.1 μg/ml tunicamycin (Sigma-Aldrich). Cells were checked every day for colony formation and cell number, and viability was assessed through Trypan blue staining. At the respective drug-treatment endpoints, cells were fixed with 70% ethanol for 5 min at room temperature and stained with Giemsa (Sigma-Aldrich). Colonies (defined as clusters of ≥50 cells) were counted with the use of a microscope (3× magnification).38
Statistical Analyses
Sample-size power was calculated with OpenEpi software (see Web Resources); a population size of 1,000,000 individuals and a hypothesized percent frequency of outcome factor (SEC23B-variant-positive status) of 2% ± 5% were used. The sample size (n) was predicted for various confidence levels (80%–99.99%).
Standardized incidence rate (SIR) for thyroid cancer is a summary ratio that allows comparison of incidence rates of thyroid cancer between affected (variant-positive) individuals and the general population. The expected number of thyroid cancers was obtained with the use of age-specific Surveillance Epidemiology and End Results (SEER) incidence rates from 2008 to 2012. There were 19 categories based on two genders and 19 age groups. Incidence was assumed to be zero for categories where statistics were not provided. Years of observation time per person (person-years of observation) were calculated from the date of birth to the date of cancer diagnosis for individuals who developed thyroid cancer or to the date of most recent information for individuals without cancer. We calculated the expected number of cancers by multiplying SEER incidence rates in each of the 19 categories by person-years of observation in each category. OpenEpi software was used for calculating SIR. The 95% confidence interval (CI) and corresponding p value were calculated with the mid-p exact test. A SIR > 1.0 implies that the incidence rate is greater for the population of interest than for the standard population. For analyses between affected population groups, we used 2 × 2 tables to calculate the odds ratio (OR) and 95% CI and used the mid-p exact test to calculate corresponding p values.
Experimental data between wild-type and mutant cell lines are given as means ± SEM, and n corresponds to the number of experiments performed. Student’s t test was used for significance testing as indicated in the figure legends. All statistical tests were two sided, and p values ≤ 0.05 were deemed significant.
Results
Exome Sequencing and Targeted Genotyping Identifies SEC23B as a Candidate Gene in a CS-Affected Family with Predominant Thyroid Cancer
We identified a multi-generation CS-affected family (family 616) presenting with different types of CS-related cancers, particularly thyroid cancer, across four generations. The family pedigree suggests autosomal-dominant inheritance with complete penetrance (Figure 1). The proband (III-2) is a 60-year-old female who presented at our CS clinic with follicular variant papillary thyroid cancer (FvPTC; diagnosed at age 43), goiter, breast fibroadenoma and papilloma, typical breast ductal hyperplasia, fibrocystic breast disease, trichilemmoma, papillomatous papules of the mucosa, gastrointestinal polyps, and uterine fibroids. The individual had a high CC score of 16, but importantly, analyses for PTEN intragenic mutations and deletions revealed a wild-type sequence. This suggested a high phenotypic burden independent of PTEN, and subsequent testing also showed no germline mutations or deletions in SDHB, SDHC, or SDHD and the absence of KLLN hypermethylation. We therefore recruited the proband for our whole-exome sequencing efforts to identify previously unidentified candidate genes in mutation-negative CS individuals with high CC scores. Exome sequencing of germline genomic DNA derived from III-2 achieved a mean coverage of 96.3×, and >90% of the targeted regions were covered by at least ten reads. Filtering the gene variants through ANNOVAR22 and Condel23 and subsequent manual curation through the IGV27, 28 retained 44 potentially deleterious gene variants, of which 39 were validated by Sanger sequencing (Table S2).
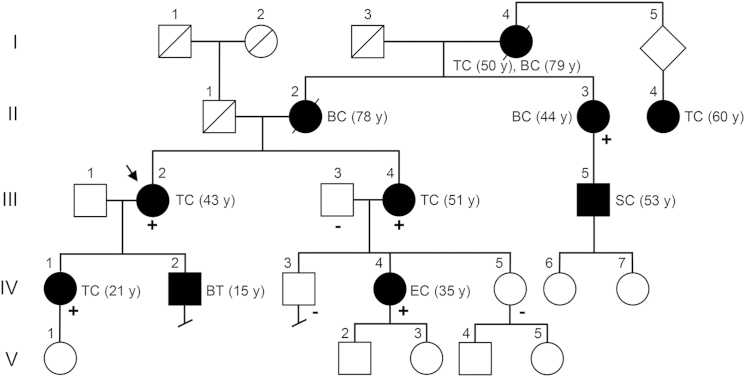
Figure 1.
Pedigree of CS-Affected Family 616
The pedigree of CS-affected family 616 is shown with the presence of wild-type (−) or c.1781T>G (p.Val594Gly) mutant (+) SEC23B alleles. Symbols are as follows: square, male; circle, female; black fill, affected; slash, deceased. The proband (III-2) for whom whole-exome sequencing was performed is indicated by an arrow. Roman numerals represent generation number, and Arabic numerals represent individuals within each generation. Affected members show various types of CS-related cancers, including thyroid (II-4, III-2, III-4, and IV-1), breast (II-2 and II-3), endometrial (IV-4), skin (III-5), and both thyroid and breast (I-4) cancers. Age at diagnosis is indicated between parentheses. Abbreviations are as follows: TC, thyroid cancer; BT, benign thyroid; BC, breast cancer; EC, endometrial cancer; SC, skin cancer.
To identify candidate cancer-predisposing CS-related genes segregating in the proband’s family, we applied Sanger sequencing for the 39 validated gene variants in seven other family members with available DNA (II-3, III-3, III-4, IV-1, IV-3, IV-4, and IV-5). Four (II-3, III-4, IV-1, and IV-4) met CS diagnostic criteria and presented with different cancer types, whereas the remaining three (III-3, IV-3, and IV-5) had no apparent phenotypes. Upon variant filtration (Table S2), all affected members were found to share three genes with heterozygous missense variants: C16orf72 (c.253T>C [p.Ser85Pro]), PTPN2 (c.1204G>A [p.Ala402Thr]), and SEC23B (c.1781T>G [p.Val594Gly]). Notably, no variants were noted in PTEN, SDHB, SDHC, SDHD, KLLN, PIK3CA, or AKT1. The three variants in SEC23B, C16orf72, and PTPN2 have not been reported in public databases (dbSNP137, 1000 Genomes, or ESP6500). We examined in silico the predicted functional impact of these prioritized variants and noted a higher index of pathogenicity for the SEC23B variant according to multiple mutation prediction databases and multi-species evolutionary amino acid residue conservation (Table 1). We next inspected the protein status of the prioritized genes in tissues of interest through the Human Protein Atlas.39, 40 We found that only PTPN2 and SEC23B are expressed in thyroid, breast, and/or endometrial normal and cancer tissues (organs affected in family 616). However, given that the PTPN2 variant was predicted as the least damaging and annotated as a “polymorphism” by MutationTaster, the combined in silico data indicate that the SEC23B variant is the most critical germline variant in family 616 (Figure S1).
Table 1.
Characteristics of the Three Candidate Variants Identified in Affected Members of Family 616
|
Genes |
|||
|---|---|---|---|
| SEC23B | C16orf72 | PTPN2 | |
| Genomic positiona | chr20: 18,529,290 | chr16: 9,186,804 | chr18: 12,794,321 |
| DNA variant | c.1781T>G | c.253T>C | c.1204G>A |
| Protein alteration | p.Val594Gly | p.Ser85Pro | p.Ala402Thr |
| Predicted Effect of Missense Variants | |||
| Condelb | deleterious | deleterious | deleterious |
| MutationTaster | disease causing | disease causing | polymorphism |
| MutPred g score (>0.5) | 0.785 | 0.525 | 0.578 |
| CADD scorec | 22.2 | 23.2 | 9.28 |
| Evolutionary Conservation and Allele Frequency | |||
| PhyloPd | 4.572 | 2.882 | 1.282 |
| PhastConse | 1 | 1 | 0.994 |
| NHLBI ESPf MAF | G = 0, T = 13,006 | C = 0, T = 12,994 | T = 0, C = 13,006 |
| Tissue-Specific Protein Levelsg | |||
| Thyroid | + | − | + |
| Breast | + | − | + |
| Endometrium | + | − | + |
Genomic positions correspond to the UCSC hg19 reference assembly.
Condel combines multiple prediction software algorithms (SIFT, PolyPhen-2, and MutationAssessor).
CADD-scaled C score ≥ 20 indicates variants within the top 1% of likely deleterious variants across the human genome.
PhyloP scores range between −14 and +6; conserved sites have positive scores.
PhastCons scores range between 0 and 1; values closer to 1 have a higher probability of nucleotide conservation.
NHLBI Exome Sequencing Project (ESP) Exome Variant Server (see Web Resources) v.0.0.28 (accessed May 9, 2015).
Data analyzed from the Human Protein Atlas for immunohistochemically stained normal and cancer tissues.
Germline Heterozygous SEC23B Variants Exist in Unrelated CS Probands with Thyroid Cancer
We next sought to investigate whether germline variants in SEC23B and the other two prioritized genes exist in other CS individuals. We selected 96 CS and CSL probands with either a diagnosis of thyroid cancer or signs of other cancers observed in family 616, namely breast and endometrial carcinomas (Table S3). This sample size gave us >95% power to detect a 2% prevalence of variants. We directly sequenced all exons of SEC23B, PTPN2, and C16orf72. We found deleterious germline heterozygous SEC23B variants in three probands (3.1%). Papillary thyroid cancer (PTC) was found in all three individuals, one of whom showed the follicular variant on histology. Interestingly, two individuals had the same missense SEC23B variant (c.490G>T [p.Val164Leu]) affecting a highly evolutionarily conserved residue within the SEC23B trunk domain,41 and all three variants we found are rare in the general population and predicted to be damaging (Table 2). In support of our in silico gene prioritization, no germline variants were found in C16orf72 or PTPN2 in the 96 unrelated CS and CSL probands.
Table 2.
Genotype and Clinical Phenotype of Known Mutation-Negative CS and/or CSL Individuals with Germline SEC23B Variants
|
Probands |
|||
|---|---|---|---|
| CCF02565 | CCF05664 | CCF04372 | |
| Age | 59 years | 69 years | 51 years |
| Sex | female | female | female |
| CC score | 7 | 11 | 14 |
| Clinical features | FvPTC (age 52 years), macrocephaly | PTC (age 45 years), ductal carcinoma in situ, atypical ductal hyperplasia, fibrocystic breast disease, hemangioma, macrocephaly | PTC (age 45 years), goiter, Hashimoto thyroiditis, macrocephaly |
| SEC23B variant | c.490G>T (p.Val164Leu), rs36023150 |
c.490G>T (p.Val164Leu), rs36023150 |
c.1512T>C (p.=), near splice, rs138198461 |
| NHLBI ESP MAF | T = 97, G = 12,909 (0.0075) | T = 97, G = 12,909 (0.0075) | C = 52, T = 12,954 (0.0040) |
| SIFT | damaging (0.01) | damaging (0.01) | – |
| PolyPhen-2 | possibly damaging (0.87) | possibly damaging (0.87) | – |
| MutationTaster | disease causing | disease causing | disease causing (acceptor lost) |
| PhyloPa | 5.615 | 5.615 | – |
| PhastConsb | 1 | 1 | – |
Abbreviations are as follows: FvPTC, follicular variant papillary thyroid cancer; PTC, papillary thyroid cancer.
PhyloP scores range between −14 and +6; conserved sites have positive scores.
PhastCons scores range between 0 and 1; values closer to 1 have a higher probability of nucleotide conservation.
Germline Heterozygous SEC23B Variants Are Enriched in Apparently Sporadic Thyroid Cancer and Associate with Early Age of Onset
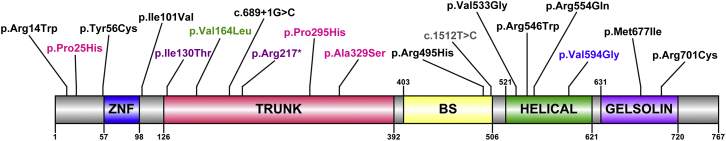
To determine whether SEC23B is associated with apparently sporadic cancers, we next analyzed the spectrum and frequencies of germline variants in three TCGA CS-component cancers (also observed in family 616): papillary thyroid (n = 494), breast invasive (n = 222), and uterine corpus endometrioid (n = 156) carcinomas. Indeed, we found germline heterozygous variants in all three studied cancer types, and the highest frequency (4%) was observed in thyroid cancer. Furthermore, we observed an overrepresentation of unique SEC23B variants in thyroid cancer (Table 3). All variants we found are predicted to be deleterious and have various protein structural effects (Table S4). For affirmation, we also inspected C16orf72 and PTPN2 and observed the absence of mutations or much lower mutation frequencies in the three cancer types (Table S5). Although we used a small sample size, we calculated the age-adjusted SIR of thyroid cancer in SEC23B-variant-positive TCGA individuals (n = 19) and found that it is significantly higher than in the US general population (SIR = 242.6; 95% CI = 150.4–371.8; p = 1.0 × 10−11). Relatedly, the median age at diagnosis of thyroid cancer is younger for SEC23B-variant-positive individuals, especially for those with unique variants, than for the entire TCGA thyroid cancer cohort (36 versus 46 years, p = 0.025). One such individual who was diagnosed with PTC at age 21 had a parent with thyroid cancer. Lastly, three unrelated individuals harbored the same SEC23B variant (c.1598T>G [p.Val533Gly]) affecting the helical SEC23B domain, where the p.Val594Gly variant from family 616 also occurs. The spectrum of germline SEC23B variants in CS and TCGA CS-related cancers is depicted in Figure 2.42
Table 3.
Characteristics of Germline SEC23B Variants in TCGA Thyroid, Breast, and Endometrial Cancer Datasets
| Cancer Datasets | Variants | NHLBI ESPaMAF | SIFT | PolyPhen-2 | MutationTaster |
Conservation |
|
|---|---|---|---|---|---|---|---|
| Phylob | PhastConsc | ||||||
| THCA (n = 494) | c.1661G>A (p.Arg554Gln) | 0 | damaging (0.02) | probably damaging (0.998) | disease causing | 5.365 | 1 |
| c.1598T>G (p.Val533Gly) | 0 | damaging (0) | possibly damaging (0.621) | disease causing | 4.497 | 1 | |
| c.167A>G (p.Tyr56Cys) | 0 | damaging (0) | possibly damaging (0.752) | disease causing | 4.798 | 1 | |
| c.301A>G (p.Ile101Val) | 0 | damaging (0.04) | possibly damaging (0.643) | disease causing | 4.760 | 1 | |
| c.689+1G>C | 0 | – | – | disease causing | 5.047 | 1 | |
| c.2101C>T (p.Arg701Cys) | 0 | damaging (0) | probably damaging (1.000) | disease causing | 2.648 | 0.988 | |
| c.1636C>T (p.Arg546Trp) | T = 1, C = 13,005 (0.000077) | damaging (0) | probably damaging (1.000) | disease causing | 1.987 | 1 | |
| c.2031G>A (p.Met677Ile) | A = 2, G = 13,004 (0.000154) | damaging (0.02) | benign (0.014) | disease causing | 5.584 | 1 | |
| c.40C>T (p.Arg14Trp) | T = 3, C = 13,003 (0.000231) | damaging (0.01) | probably damaging (0.999) | disease causing | 2.132 | 1 | |
| c.1484G>A (p.Arg495His) | A = 95, G = 12,911 (0.007358) | tolerated (0.12) | probably damaging (0.961) | disease causing | 5.579 | 1 | |
| c.490G>T (p.Val164Leu) | T = 97, G = 12,909 (0.0075) | damaging (0.01) | possibly damaging (0.870) | disease causing | 5.615 | 1 | |
| BRCA (n = 222) | c.884C>A (p.Pro295His) | 0 | damaging (0) | probably damaging (0.983) | disease causing | 5.778 | 1 |
| c.985G>T (p.Ala329Ser) | T = 4, G = 13,002 (0.000307) | damaging (0.04) | possibly damaging (0.601) | disease causing | 5.591 | 1 | |
| c.74C>A (p.Pro25His) | A = 5, C = 13,001 (0.000385) | damaging (0) | probably damaging (1.000) | disease causing | 5.701 | 1 | |
| c.490G>T (p.Val164Leu) | T = 97, G = 12,909 (0.0075) | damaging (0.01) | possibly damaging (0.870) | disease causing | 5.615 | 1 | |
| UCEC (n = 156) | c.389T>C (p.Ile130Thr) | 0 | damaging (0.05) | benign (0.006) | disease causing | 4.638 | 1 |
| c.649C>T (p.Arg217∗) | T = 1, C = 13,005 (0.000077) | – | – | disease causing | −0.056 | 0.031 | |
| c.490G>T (p.Val164Leu) | T = 97, G = 12,909 (0.0075) | damaging (0.01) | possibly damaging (0.870) | disease causing | 5.615 | 1 | |
NHLBI Exome Sequencing Project (ESP) Exome Variant Server v.0.0.28 (accessed May 9, 2015).
PhyloP scores range between −14 and +6; conserved sites have positive scores.
PhastCons scores range between 0 and 1; values closer to 1 have a higher probability of nucleotide conservation.
Figure 2.
Germline SEC23B Mutation Spectra in CS and Apparently Sporadic CS-Related Cancers from TCGA
A schematic of SEC23B and its functional domains is shown; Arabic numerals correspond to amino acid number. The amino acid positions corresponding to germline heterozygous variants are indicated for variants identified in CS-affected family 616 (blue), an unrelated CS proband (gray), TCGA apparently sporadic thyroid cancer (black), breast cancer (pink), endometrial cancer (purple), and a shared variant observed in all three cancer types and in CS (green). Abbreviations are as follows: ZNF, zinc finger domain; BS, beta-sandwich.
Our findings indicate that both unique and previously reported (0.00008 ≤ MAF ≤ 0.008) germline SEC23B variants exist in CS individuals and in the studied individuals with sporadic cancer. This prompted us to investigate the frequency of heterozygous SEC23B variants in individuals with no reported cancer diagnoses. We analyzed the NHLBI exomes representing 2,203 African-American and 4,300 European-American unrelated individuals, totaling 13,006 chromosomes. We found deleterious SEC23B variants in 155 (1.2%; MAF ≤ 0.008) or 58 (0.45%; MAF ≤ 0.0004) chromosomes (Table S6). In comparison, the studied TCGA populations with apparently sporadic cancer show deleterious SEC23B variants in 29 (1.7%; MAF ≤ 0.008) or 16 (0.93%; MAF ≤ 0.0004) out of 1,744 chromosomes. Interestingly, this comparison yields a significantly higher OR for thyroid cancer in TCGA populations than in NHLBI populations, especially for variants with a MAF ≤ 0.0004 (OR = 2.5; 95% CI = 1.3–4.7; p = 0.0116) (Table S7).
Functional Characterization of p.Val594Gly in Family 616 Reveals a Tumorigenic Cellular, Molecular, and Biochemical Phenotype
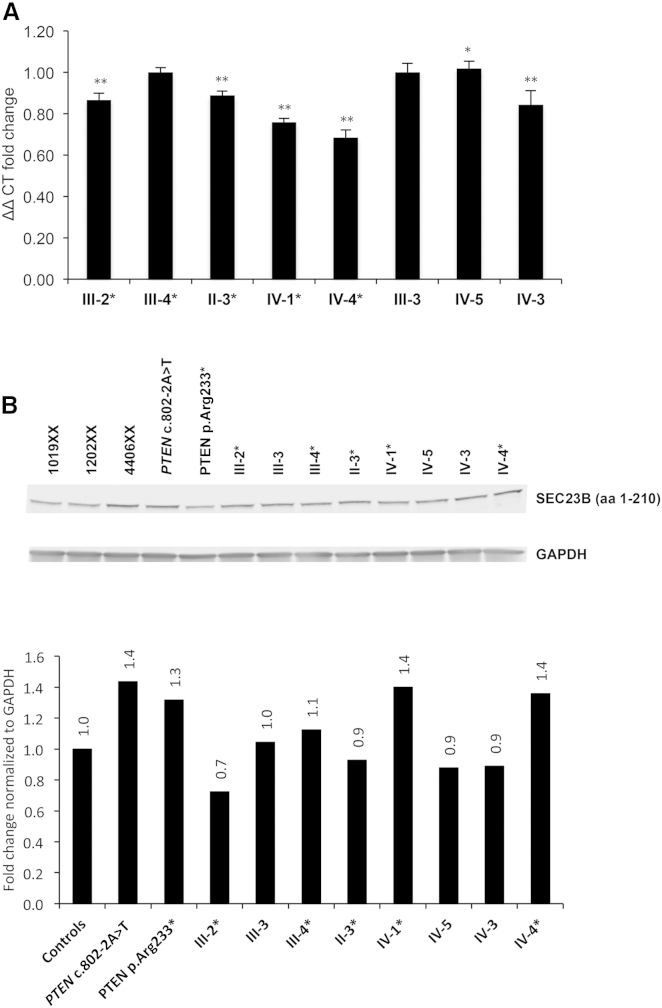
SEC23B is a component of COPII vesicles for anterograde ER-to-Golgi transport.43, 44 Recessive loss-of-function mutations cause congenital dyserythropoietic anemia type II (CDAII),45, 46 resulting in decreased SEC23B levels.45, 47, 48 Available clinical laboratory results from a subset of the available heterozygous SEC23B-variant-positive CS individuals were not suggestive of a classic CDAII phenotype, and reports signed off as “normal” for most of these individuals (Table S8). Also, lymphoblastoid cells derived from family 616 showed no differences in SEC23B transcript and protein levels between variant carriers and wild-type individuals (Figure 3), suggesting that a probable change in function rather than reduced SEC23B levels (loss of function) is the likely mechanism in CS.
Figure 3.
Lymphoblastoid Cells Derived from Family 616 Show No Differences in SEC23B Transcript and Protein Levels between p.Val594Gly Carriers and Wild-Type Individuals
(A) qRT-PCR expression showed no notable trend in SEC23B transcript expression between wild-type individuals and variant carriers. Values are normalized to unaffected individual III-3 and represent three technical replicates for which data represent mean values ± SEM. ∗∗p < 0.01, ∗p < 0.05 (two-sided Student’s t test).
(B) SEC23B levels and quantification by western blot. PTEN-mutation-positive lymphoblastoid cells (LCLs) from other CS individuals were also used as internal exploratory controls. 1019XX, 1202XX, and 4406XX represent healthy control LCLs unrelated to the pedigree.
SEC23B-mutation carriers from the pedigree are indicated by asterisks in each panel. “aa” refers to the amino acids targeted by the SEC23B antibody.
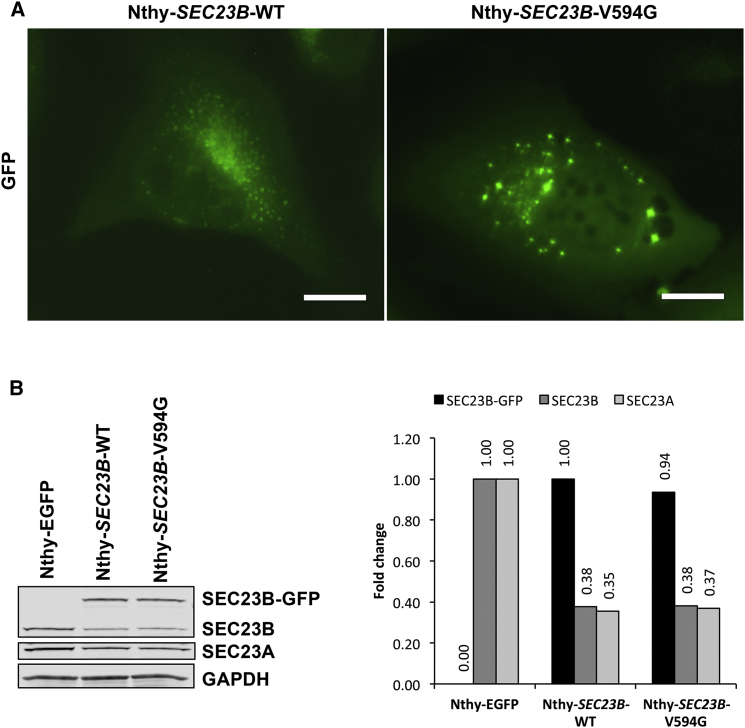
We then sought to functionally characterize the unique deleterious SEC23B variant (c.1781T>G [p.Val594Gly]) found in family 616. We transiently transfected HEK293T cells with wild-type and p.Val594Gly SEC23B. Mutant cells revealed aberrant aggregation of SEC23B and significantly increased cell migration and upregulation of epithelial-to-mesenchymal transition (EMT) genes, suggesting a cancer-relevant phenotypic effect (Figure S2). Notably, the p.Val594Gly variant occurs between the alpha-N and alpha-M regions within the SEC23B helical domain critical for binding to SAR1,49 a GTPase activated by SEC23 and another component of the inner coat of COPII vesicles. We speculated that the observed alteration might affect binding of SEC23B to SAR1A and confirmed this through immunoprecipitation of tagged SEC23B and SAR1A in 293T-SEC23B-WT and 293T-SEC23B-V594G cells (Figure S3). Pertinent to the cancer type (i.e., thyroid) enriched in family 616 and TCGA, we then generated stable normal thyroid follicule epithelial50 cell lines expressing wild-type or p.Val594Gly SEC23B (Nthy-SEC23B-WT or Nthy-SEC23B-V594G, respectively). We validated that compared to Nthy-SEC23B-WT and Nthy-SEC23B-E109K cells (the latter of which represents one of two CDAII founder mutations accounting for ∼50% of reported cases51, 52), the Nthy-SEC23B-V594G cells associated with the CS-affected family showed aberrant SEC23B aggregation (Figure 4A and Figure S4). The Nthy-SEC23B-E109K cells lacked a normal ER expression pattern, supporting the loss of altered protein in anemia. Relatedly, western blot analysis revealed similar levels of SEC23B and paralog SEC23A in wild-type and mutant Nthy-SEC23B-V594G cells, adding to the evidence that the mutation does not result in loss of function of SEC23B and does not lead to concomitant compensation by SEC23A (Figure 4B).53
Figure 4.
Cellular and Biochemical Characterization of Family 616 Variant p.Val594Gly in a Normal Thyroid Cell Line
(A) Normal thyroid Nthy-ori 3-1 cells were stably transduced with wild-type and mutant c.1781T>G (p.Val594Gly) SEC23B fused with GFP. Wild-type cells (Nthy-SEC23B-WT) showed SEC23B localization typical of ER proteins, whereas a subset of mutant cells (Nthy-SEC23B-V594G) showed aberrant aggregation of SEC23B. Scale bars represent 20 μm.
(B) Western blot and densitometric analysis of overexpressed SEC23B, endogenous SEC23B and paralog SEC23A, and GAPDH (loading control) from stable cell lines. Wild-type and mutant Nthy-SEC23B-V594G cells showed similar levels of SEC23B and SEC23A. The Nthy-EGFP cell line was used as a negative control to indicate baseline levels of SEC23B and SEC23A in the cell line (no SEC23B overexpression in these cells). Exogenous SEC23B-GFP normalized to the wild-type, and endogenous SEC23B and SEC23A normalized to levels in the Nthy-EGFP cell line.
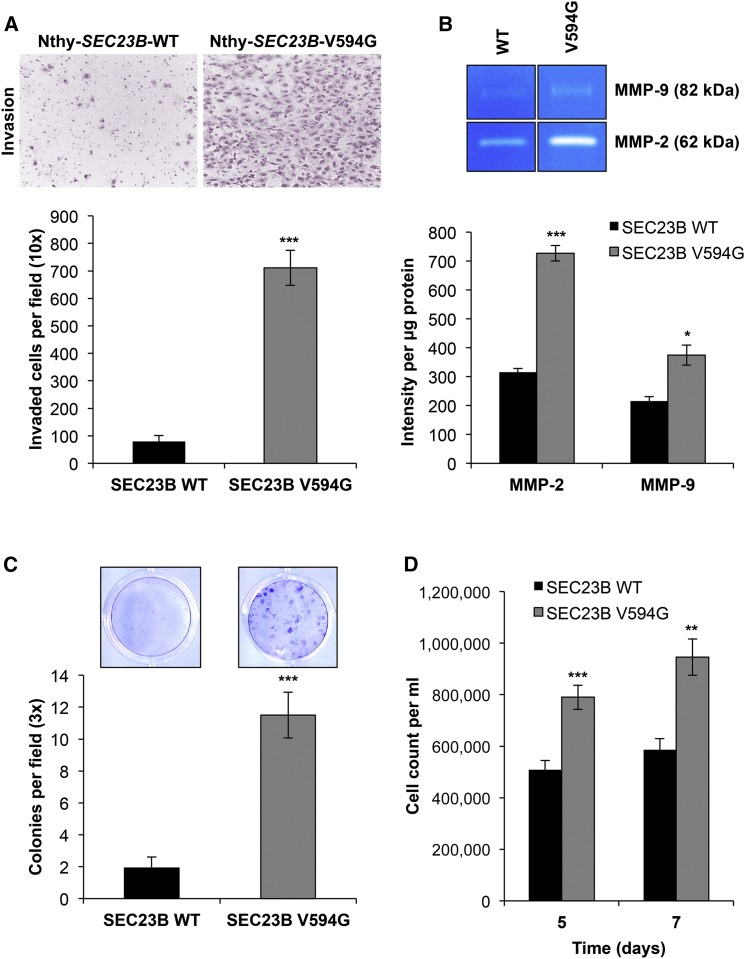
To our surprise, Nthy-SEC23B-WT and Nthy-SEC23B-V594G cells showed no differences in growth, viability, or migration ability (Figure S5). We hypothesized that another aspect of the EMT phenotype might be affected in normal thyroid cells, and so we turned our attention to investigating other EMT-related manifestations (or phenotypes). Indeed, Transwell invasion assays revealed a significantly higher (9-fold) increase in Matrigel invasion (p = 2.31 × 10−7) in Nthy-SEC23B-V594G cells (Figure 5A). We then demonstrated through gelatin zymography that these mutant cells showed increased activity of MMP-2 and MMP-9 (matrix-digesting metalloproteinases) in the supernatant (Figure 5B), thus explaining the invasive potential of these cells.
Figure 5.
The SEC23B c.1781T>G (p.Val594Gly) Variant Confers Neoplastic Phenotypes upon ER Stress
(A) Representative images of invaded Nthy-SEC23B-WT (left) and Nthy-SEC23B-V594G (right) cells in a Transwell assay after overnight serum starvation. Invaded cells were counted after 24 hr in quadruplicate fields of view in triplicate membranes. Data represent mean values ± SEM. ∗∗∗p < 0.001 (two-sided Student’s t test).
(B) Gelatin zymography was performed with conditioned media from wild-type and mutant cells and showed more MMP-2 and MMP-9 activity in supernatant originating from Nthy-SEC23B-V594G (right lanes) than in supernatant from Nthy-SEC23B-WT (left lanes) cells. Data represent triplicate lanes with band intensity normalized to protein mass. Data represent mean values ± SEM. ∗∗∗p < 0.001, ∗p < 0.05 (two-sided Student’s t test).
(C) Wild-type and mutant cells were treated with 1 μM thapsigargin (TG) for the induction of ER stress. Large colonies of cells expressing altered SEC23B formed, after which both Nthy-SEC23B-WT and Nthy-SEC23B-V594G cells were fixed and stained with Giemsa, and colonies were counted. Stained colonies represent cells 14 days after treatment, and a colony is standardly defined as a cluster of ≥50 cells. Data shown represent three independent experiments done in triplicate for each genotype. Data represent mean values ± SEM. ∗∗∗p < 0.001 (two-sided Student’s t test).
(D) Treatment of wild-type and mutant cells with 0.1 μg/ml tunicamycin (TM), another ER-stress-inducing agent, resulted in more growth of mutant cells than of wild-type cells. Data shown represent two independent experiments done in triplicate for each genotype at each time point. Data represent pooled mean values ± SEM. ∗∗∗p < 0.001, ∗∗p < 0.01 (two-sided Student’s t test).
We continued to examine other cellular phenotypes linked to carcinogenesis. Given that loss of SEC23B in mice causes ER stress and degeneration of secretory organs,36 we hypothesized that heterozygous SEC23B variants might prime mutant cells to respond differently to ER-stress stimuli. We first treated cells with thapsigargin, an ER Ca2+ ATPase inhibitor conventionally used to induce ER stress experimentally. Previous studies have reported that ER stress can induce an EMT phenotype in thyroid and lung alveolar epithelial cells.54, 55, 56 Consistent with these studies,54, 55, 56 starting at 6 hr after treatment with thapsigargin, both wild-type and mutant cells showed a cellular and molecular phenotype consistent with EMT (Figure S6). Intriguingly, cellular phenotypic differences between wild-type and Nthy-SEC23B-V594G cells became evident only after chronic exposure to thapsigargin (at ∼14 days) in that mutant cells formed significantly larger colonies (p = 1.84 × 10−5) that survived for up to 36 days after treatment (Figure 5C and Figure S7). In support of these findings, treatment with tunicamycin, another standard ER-stress inducer (inhibitor of protein N-linked glycosylation),57 also resulted in the formation of distinct colonies and conferred an apparent growth advantage at lower doses in mutant cells (Figure 5D). Overall, our experimental findings demonstrate examples of how a SEC23B germline variant could account for increased susceptibility to cancer, particularly thyroid cancer.
Discussion
Our observations reveal that a subset of CS and CSL individuals without germline mutations in known genes might be accounted for by germline variants in SEC23B. Of the three SEC23B variants we identified in four unrelated CS and/or CSL individuals, the pathogenic c.1781T>G (p.Val594Gly) variant related to family 616 has never been described before; our functional analyses corroborate the pathogenicity of this missense mutation. Structurally, p.Val594Gly occurs between the alpha-N and alpha-M regions within the SEC23B helical domain critical for binding to SAR1, a GTPase activated by SEC23 and another component of the inner coat of COPII vesicles. Interestingly, the p.Val164Leu variant present in two other unrelated CS and/or CSL individuals occurs within a different protein domain, the SEC23 trunk domain. This region forms the dimer interface between SEC23 and SEC24 and also contacts SAR1,41 suggesting a possible common downstream effect. This variant has been reported in ESP6500 with a MAF of 0.0075 (97/12,909). Therefore, although we cannot ascertain causation, we bear in mind that cancer is a common phenotypic manifestation even in the general population. For example, according to SEER, the risk of thyroid cancer in the general population is 1%, that of female breast cancer is 12%, and that of endometrial cancer is 2.6%.13 Hence, it is possible that this SEC23B p.Val164Leu variant might be predisposing these individuals to cancer even in the general population.
It is also worth mentioning that SEC23B p.Val164Leu is reminiscent of two SDHD variants (c.34G>A [p.Gly12Ser] and c.149A>G [p.His50Arg]) with well-established functional roles in CS and CSL. Although c.34G>A and c.149A>G are reported in ESP6500 with a MAF of 0.0080 (103/12,893) and 0.0063 (82/12,914), respectively, we have shown that these two variants result in increased reactive oxygen species (via PHD) and upregulation of p-AKT and p-ERK1/2 (mimicking loss of function of PTEN).9 Furthermore, they also destabilize p53 by altering the NAD/FAD ratio, which leads to decreased NQ01-p53 interaction with consequent non-MDM2 degradation of p53.10 More recently, we also elucidated that these same SDHD variants lead to nuclear mislocalization and oxidation of PTEN in CS-related thyroid cancer.58 Therefore, although SEC23B might be a pertinent cancer-predisposing gene, as demonstrated by our functional evidence of the c.1781T>G (p.Val594Gly) variant, the relevance of the c.490G>T (p.Val164Leu) variant would require similar experimental interrogation for more conclusive evidence, although we believe that the latter could be a low- to moderate-penetrance allele. Lastly, because ESP6500 represents a set of population controls, it is plausible that some individuals might have unrecognized CS and CSL, given the subtlety of some of the clinical features and the existence of phenotypes often observed in the general population, such as cancer and non-specific skin features.
We also found heterozygous germline SEC23B variants in apparently sporadic breast, thyroid, and endometrial cancers in TCGA. Notably, these variants were overrepresented in individuals with thyroid cancer and enriched with unique SEC23B variants predicted to be deleterious and absent from public databases, suggesting a functional role in sporadic thyroid carcinogenesis. There is no overlap of mutation spectra between CDAII52 and CS. However, five TCGA individuals with cancer harbored germline heterozygous SEC23B variants (c.40C>T [p.Arg14Trp], c.649C>T [p.Arg217∗], c.2101C>T [p.Arg701Cys], c.74C>A [p.Pro25His], and c.689+1G>C) known to be associated with CDAII in the homozygous or compound-heterozygous setting. Because we cannot ascertain anemia-related clinical phenotypes unrelated to cancer in these individuals, such variants warrant further investigation. Interestingly, irrespective of SEC23B-mutation status, an agnostic search in the Catalogue of Somatic Mutations in Cancer (COSMIC) showed that 625/828 (∼75%) of apparently sporadic cancers of different types include overexpression of SEC23B transcripts (Z score values > 2) and that 84/828 (∼10%) show high-level amplifications (gain of copy number), suggesting that at least in a subset of cancers, SEC23B could indeed be playing a previously unappreciated role in carcinogenesis at large.
Given the role of SEC23B, we speculate that because of the high demand for protein synthesis and trafficking and, hence, increased sensitivity to perturbations in ER function, germline SEC23B variants associate with cancers in so-called professional secretory tissues (in this study, thyroid, breast, and endometrium). Our functional data also suggest that p.Val594Gly is a change-of-function substitution that favors tumorigenesis even, or especially, under stressful micro-environmental conditions. Cells respond to ER stress by activating the unfolded protein response (UPR), a signaling network that allows the recovery of ER function by upregulating adaptive factors, such as chaperone proteins and transcription factors, that regulate protein synthesis, growth arrest, and ER biogenesis among various pro-survival responses.59 Cell fate is largely determined by the intensity and duration of the stress stimuli, and failure to adapt to ER stress ultimately leads to the activation of apoptotic pathways. Increasing evidence, however, suggests that cancer cells hijack or are addicted to ER-stress-induced responses as a survival mechanism and thus often confer resistance to anti-cancer therapies.60, 61, 62, 63, 64, 65
Mouse models completely deficient of SEC23B die shortly after birth.36 Interestingly, these mice do not have anemia but rather suffer from UPR-induced degeneration of professional secretory tissues that seem to highly depend on SEC23B for efficient cargo transport and secretion. More recently, it was reported that mice with hematopoietic compartment localized deficiency of SEC23B also show no red blood cell phenotypes.66 The difference in phenotypes observed between humans and mice was attributed to the existence of hypomorphic mutations in humans and a potential species-specific overlap of function with the SEC23A paralog. Interestingly, Korpal and colleagues agnostically identified that direct targeting of SEC23A by miR200s promotes metastatic lung colonization in mice by inhibiting the secretion of metastasis-suppressive proteins and promoting the mesenchymal-to-epithelial transition (MET).67 The appearance of large epithelial colonies in our mutant Nthy-SEC23B-V594G cells after ER stress is suggestive of a phenotype favoring cell colonization. Whether this is conducive to primary tumor formation or metastatic cell seeding has yet to be determined. Moreover, increased MMP-2 and MMP-9 activity from the cell supernatant hints at the possibility that protein-trafficking alterations modulate the cell-derived secretome similarly to the newly discovered SEC23A role in trafficking metastasis-regulating proteins.67 Furthermore, the different migration patterns between Nthy cells and HEK293T cells transfected with our mutant SEC23B might lend a clue. Although we could attribute differences to varying cellular contexts, HEK293T cells are immortalized with SV40 large T antigen, whereas Nthy cells are immortalized with SV40, where the origin of replication is absent. One way or the other, we speculate that the suspected heterozygous change-of-function SEC23B mutations in a cancer context could override the existence of wild-type SEC23A and SEC23B, at least in thyroid, breast, and endometrial cancers, potentially through ER stress and UPR “addiction.”
However, the precise mechanism leading to neoplastic transformation warrants further investigation. There is indeed increasing evidence that monoallelic and biallelic mutations in a single gene can lead to completely different phenotypes.5, 68, 69 Relatedly, there are currently no reports of cancer predisposition in the obligate heterozygous parents of individuals with CDAII. One possibility is that the carried loss-of-function CDAII-associated mutations result in a less severe hypomorphic phenotype even in the heterozygous state, whereas we speculate that cancer-promoting heterozygous variants do not affect protein stability and result in change-of-function effects. The other possibility is that parents of CDAII individuals, especially if the latter are pediatric, might not be followed up long enough for the existence of cancer to be ascertained. This is confounded by the fact that CDAII is a rare disorder with a global series of only 205 affected individuals (172 unrelated),52 and so it is plausible that the existence of common cancers in these families might go unnoticed. This phenomenon is paralleled by the discovery of germline heterozygous mutations in FH (MIM: 136850), encoding fumarate hydratase, in hereditary leiomyomatosis and renal cell carcinoma (HLRCC [MIM: 150800]).69 Homozygous or compound-heterozygous FH mutations cause fumarase deficiency (FMRD [MIM: 606812]), a lethal neuropathy, cardiopathy, and/or hepatopathy in infancy or early childhood.68, 70 The heterozygous parents of the individuals with these rare diseases are not documented to have had cancer. Similarly, heterozygous SDHA, SDHB, SDHC, and SDHD mutations were described to cause endocrine neoplasias71, 72, 73 only 13 years ago, whereas germline homozygous or compound-heterozygous mutations in SDHA, SDHB, SDHC, and SDHD (any one of four subunits of succinate dehydrogenase or mitochondrial complex II) have long been known to cause Leigh syndrome, a lethal neuropathy that manifests shortly after birth.74, 75 Yet, none of the parents of individuals with Leigh syndrome have been reported in the literature to have had component cancers. Quite similar to Sec23b mouse models, the existing murine models of Sdhb, Sdhd, and Fh do not mimic the human neoplasia conditions and indeed have little to no phenotype (reviewed by Lepoutre-Lussey et al.76). It is tempting to speculate on a “common” mechanism that might explain the disparate phenotypes, namely cancer and non-neoplastic yet extremely serious phenotypes, between heterozygous and homozygous or compound-heterozygous germline mutations in seemingly disparate genes (SEC23B, SDHA, SDHB, SDHC, SDHD, and FH) and the lack of the neoplastic phenotype in any of these heterozygous mouse models.
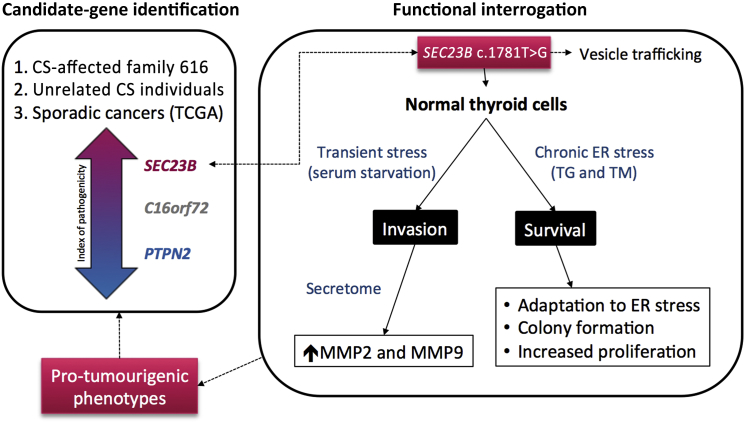
We can only speculate that tissue-specific gene-dosage thresholds and mutation-specific effects on encoded protein function might account for such differences. Notably, our observations in the context of heterozygous SEC23B mutations offer an important glimpse into how heritable gene mutations interact with the environment (e.g., an exposure leading to ER stress) to result in a particular disease phenotype (here, cancer)—and hence, genetic predisposition to cancer. Indeed, vesicular trafficking is a major contributor to carcinogenesis in that it modulates the trafficking of key proteins involved in cell polarity, motility, invasion, and cell-cell and cell-matrix interactions.77 Our finding that heterozygous SEC23B germline variants exist in CS and CSL individuals and in individuals with apparently sporadic CS-component cancer is supported by our in vitro data to indicate a role for SEC23B in cancer predisposition when germline “change-of-function” variants constitutionally exist in the heterozygous context (Figure 6).
Figure 6.
SEC23B Germline Heterozygous Variants Cause Predisposition to Hereditary and Apparently Sporadic Cancers
Abbreviations are as follows: CS, Cowden syndrome; TG, thapsigargin; TM, tunicamycin.
Acknowledgments
We are grateful to the members of family 616 and all Cowden syndrome (CS)- and CS-like-affected individuals who contributed to this study. We thank the Genomic Medicine Biorepository of the Cleveland Clinic Genomic Medicine Institute and our database and clinical research teams. We also thank Dr. Thomas LaFramboise and Darcie Seachrist (Case Western Reserve University) and Todd Romigh, Charissa Petersen, and Jessica Altemus (Cleveland Clinic Genomic Medicine Institute) for technical advice and helpful discussions. This study was funded in part by the National Cancer Institute (P01CA124570 and R01CA118989), American Cancer Society (RPG-02-151-01-CCE and Clinical Research Professorship), Breast Cancer Research Foundation, William Randolph Hearst Foundations, and Doris Duke Distinguished Clinical Scientist Award (all to C.E.) and the NHLBI (R01HL094505 to B.Z.). L.Y. is an International Fulbright Science and Technology Doctoral Fellow at the Cleveland Clinic Genomic Medicine Institute and recipient of the Dr. Michael H. Fakih pre-doctoral endowed scholarship. Y.N. is a CoGEC Scholar funded in part by National Cancer Institute grant R25TCA094186. J.N. was and M.S. is an Ambrose Monell Foundation Cancer Genomic Medicine Fellow at the Cleveland Clinic Genomic Medicine Institute. C.E. is the Sondra J. and Stephen R. Hardis Chair of Cancer Genomic Medicine at the Cleveland Clinic and an American Cancer Society Clinical Research Professor.
Published: October 29, 2015
Footnotes
Supplemental Data include seven figures and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.10.001.
Accession Numbers
Exome sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number SRA: SRP059300.
Web Resources
The URLs for data presented herein are as follows:
Condel, http://bg.upf.edu/fannsdb/
Genomic Medicine Institute calculator for PTEN CC scores, http://www.lerner.ccf.org/gmi/ccscore/
Human Protein Atlas, http://www.proteinatlas.org/
MutationTaster, http://www.mutationtaster.org/
MutPred Server, http://mutpred.mutdb.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
OpenEpi, http://www.openepi.com
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org/
TCGA Data Portal, https://tcga-data.nci.nih.gov/tcga/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Marsh D.J., Coulon V., Lunetta K.L., Rocca-Serra P., Dahia P.L., Zheng Z., Liaw D., Caron S., Duboué B., Lin A.Y. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum. Mol. Genet. 1998;7:507–515. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- 2.Orloff M.S., Eng C. Genetic and phenotypic heterogeneity in the PTEN hamartoma tumour syndrome. Oncogene. 2008;27:5387–5397. doi: 10.1038/onc.2008.237. [DOI] [PubMed] [Google Scholar]
- 3.Nelen M.R., Padberg G.W., Peeters E.A., Lin A.Y., van den Helm B., Frants R.R., Coulon V., Goldstein A.M., van Reen M.M., Easton D.F. Localization of the gene for Cowden disease to chromosome 10q22-23. Nat. Genet. 1996;13:114–116. doi: 10.1038/ng0596-114. [DOI] [PubMed] [Google Scholar]
- 4.Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X.P., Waite K.A., Pilarski R., Hampel H., Fernandez M.J., Bos C., Dasouki M., Feldman G.L., Greenberg L.A., Ivanovich J. Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am. J. Hum. Genet. 2003;73:404–411. doi: 10.1086/377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eng C. Will the real Cowden syndrome please stand up: revised diagnostic criteria. J. Med. Genet. 2000;37:828–830. doi: 10.1136/jmg.37.11.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pilarski R., Eng C. Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J. Med. Genet. 2004;41:323–326. doi: 10.1136/jmg.2004.018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan M.H., Mester J., Peterson C., Yang Y., Chen J.L., Rybicki L.A., Milas K., Pederson H., Remzi B., Orloff M.S., Eng C. A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am. J. Hum. Genet. 2011;88:42–56. doi: 10.1016/j.ajhg.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni Y., Zbuk K.M., Sadler T., Patocs A., Lobo G., Edelman E., Platzer P., Orloff M.S., Waite K.A., Eng C. Germline mutations and variants in the succinate dehydrogenase genes in Cowden and Cowden-like syndromes. Am. J. Hum. Genet. 2008;83:261–268. doi: 10.1016/j.ajhg.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni Y., He X., Chen J., Moline J., Mester J., Orloff M.S., Ringel M.D., Eng C. Germline SDHx variants modify breast and thyroid cancer risks in Cowden and Cowden-like syndrome via FAD/NAD-dependant destabilization of p53. Hum. Mol. Genet. 2012;21:300–310. doi: 10.1093/hmg/ddr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett K.L., Mester J., Eng C. Germline epigenetic regulation of KILLIN in Cowden and Cowden-like syndrome. JAMA. 2010;304:2724–2731. doi: 10.1001/jama.2010.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orloff M.S., He X., Peterson C., Chen F., Chen J.L., Mester J.L., Eng C. Germline PIK3CA and AKT1 mutations in Cowden and Cowden-like syndromes. Am. J. Hum. Genet. 2013;92:76–80. doi: 10.1016/j.ajhg.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan M.H., Mester J.L., Ngeow J., Rybicki L.A., Orloff M.S., Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin. Cancer Res. 2012;18:400–407. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngeow J., Mester J., Rybicki L.A., Ni Y., Milas M., Eng C. Incidence and clinical characteristics of thyroid cancer in prospective series of individuals with Cowden and Cowden-like syndrome characterized by germline PTEN, SDH, or KLLN alterations. J. Clin. Endocrinol. Metab. 2011;96:E2063–E2071. doi: 10.1210/jc.2011-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldgar D.E., Easton D.F., Cannon-Albright L.A., Skolnick M.H. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J. Natl. Cancer Inst. 1994;86:1600–1608. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- 16.Hemminki K., Vaittinen P. Familial cancers in a nationwide family cancer database: age distribution and prevalence. Eur. J. Cancer. 1999;35:1109–1117. doi: 10.1016/s0959-8049(99)00066-0. [DOI] [PubMed] [Google Scholar]
- 17.Eng C. Familial papillary thyroid cancer--many syndromes, too many genes? J. Clin. Endocrinol. Metab. 2000;85:1755–1757. doi: 10.1210/jcem.85.5.6632. [DOI] [PubMed] [Google Scholar]
- 18.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González-Pérez A., López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green R.C., Berg J.S., Grody W.W., Kalia S.S., Korf B.R., Martin C.L., McGuire A.L., Nussbaum R.L., O’Daniel J.M., Ormond K.E., American College of Medical Genetics and Genomics ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 31.Li B., Krishnan V.G., Mort M.E., Xin F., Kamati K.K., Cooper D.N., Mooney S.D., Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venselaar H., Te Beek T.A., Kuipers R.K., Hekkelman M.L., Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mancias J.D., Goldberg J. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol. Cell. 2007;26:403–414. doi: 10.1016/j.molcel.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Capes-Davis A., Theodosopoulos G., Atkin I., Drexler H.G., Kohara A., MacLeod R.A., Masters J.R., Nakamura Y., Reid Y.A., Reddel R.R., Freshney R.I. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int. J. Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 36.Tao J., Zhu M., Wang H., Afelik S., Vasievich M.P., Chen X.W., Zhu G., Jensen J., Ginsburg D., Zhang B. SEC23B is required for the maintenance of murine professional secretory tissues. Proc. Natl. Acad. Sci. USA. 2012;109:E2001–E2009. doi: 10.1073/pnas.1209207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang C.C., Park A.Y., Guan J.L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 38.Franken N.A., Rodermond H.M., Stap J., Haveman J., van Bree C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 39.Uhlén M., Björling E., Agaton C., Szigyarto C.A., Amini B., Andersen E., Andersson A.C., Angelidou P., Asplund A., Asplund C. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 41.Lederkremer G.Z., Cheng Y., Petre B.M., Vogan E., Springer S., Schekman R., Walz T., Kirchhausen T. Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc. Natl. Acad. Sci. USA. 2001;98:10704–10709. doi: 10.1073/pnas.191359398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
- 43.Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M.F., Ravazzola M., Amherdt M., Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 44.Fromme J.C., Orci L., Schekman R. Coordination of COPII vesicle trafficking by Sec23. Trends Cell Biol. 2008;18:330–336. doi: 10.1016/j.tcb.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Schwarz K., Iolascon A., Verissimo F., Trede N.S., Horsley W., Chen W., Paw B.H., Hopfner K.P., Holzmann K., Russo R. Mutations affecting the secretory COPII coat component SEC23B cause congenital dyserythropoietic anemia type II. Nat. Genet. 2009;41:936–940. doi: 10.1038/ng.405. [DOI] [PubMed] [Google Scholar]
- 46.Bianchi P., Fermo E., Vercellati C., Boschetti C., Barcellini W., Iurlo A., Marcello A.P., Righetti P.G., Zanella A. Congenital dyserythropoietic anemia type II (CDAII) is caused by mutations in the SEC23B gene. Hum. Mutat. 2009;30:1292–1298. doi: 10.1002/humu.21077. [DOI] [PubMed] [Google Scholar]
- 47.Punzo F., Bertoli-Avella A.M., Scianguetta S., Della Ragione F., Casale M., Ronzoni L., Cappellini M.D., Forni G., Oostra B.A., Perrotta S. Congenital dyserythropoietic anemia type II: molecular analysis and expression of the SEC23B gene. Orphanet J. Rare Dis. 2011;6:89. doi: 10.1186/1750-1172-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russo R., Langella C., Esposito M.R., Gambale A., Vitiello F., Vallefuoco F., Ek T., Yang E., Iolascon A. Hypomorphic mutations of SEC23B gene account for mild phenotypes of congenital dyserythropoietic anemia type II. Blood Cells Mol. Dis. 2013;51:17–21. doi: 10.1016/j.bcmd.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bi X., Corpina R.A., Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- 50.Lemoine N.R., Mayall E.S., Jones T., Sheer D., McDermid S., Kendall-Taylor P., Wynford-Thomas D. Characterisation of human thyroid epithelial cells immortalised in vitro by simian virus 40 DNA transfection. Br. J. Cancer. 1989;60:897–903. doi: 10.1038/bjc.1989.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russo R., Gambale A., Esposito M.R., Serra M.L., Troiano A., De Maggio I., Capasso M., Luzzatto L., Delaunay J., Tamary H., Iolascon A. Two founder mutations in the SEC23B gene account for the relatively high frequency of CDA II in the Italian population. Am. J. Hematol. 2011;86:727–732. doi: 10.1002/ajh.22096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russo R., Gambale A., Langella C., Andolfo I., Unal S., Iolascon A. Retrospective cohort study of 205 cases with congenital dyserythropoietic anemia type II: definition of clinical and molecular spectrum and identification of new diagnostic scores. Am. J. Hematol. 2014;89:E169–E175. doi: 10.1002/ajh.23800. [DOI] [PubMed] [Google Scholar]
- 53.Satchwell T.J., Pellegrin S., Bianchi P., Hawley B.R., Gampel A., Mordue K.E., Budnik A., Fermo E., Barcellini W., Stephens D.J. Characteristic phenotypes associated with congenital dyserythropoietic anemia (type II) manifest at different stages of erythropoiesis. Haematologica. 2013;98:1788–1796. doi: 10.3324/haematol.2013.085522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulianich L., Garbi C., Treglia A.S., Punzi D., Miele C., Raciti G.A., Beguinot F., Consiglio E., Di Jeso B. ER stress is associated with dedifferentiation and an epithelial-to-mesenchymal transition-like phenotype in PC Cl3 thyroid cells. J. Cell Sci. 2008;121:477–486. doi: 10.1242/jcs.017202. [DOI] [PubMed] [Google Scholar]
- 55.Zhong Q., Zhou B., Ann D.K., Minoo P., Liu Y., Banfalvi A., Krishnaveni M.S., Dubourd M., Demaio L., Willis B.C. Role of endoplasmic reticulum stress in epithelial-mesenchymal transition of alveolar epithelial cells: effects of misfolded surfactant protein. Am. J. Respir. Cell Mol. Biol. 2011;45:498–509. doi: 10.1165/rcmb.2010-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanjore H., Cheng D.S., Degryse A.L., Zoz D.F., Abdolrasulnia R., Lawson W.E., Blackwell T.S. Alveolar epithelial cells undergo epithelial-to-mesenchymal transition in response to endoplasmic reticulum stress. J. Biol. Chem. 2011;286:30972–30980. doi: 10.1074/jbc.M110.181164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee A.S. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem. Sci. 2001;26:504–510. doi: 10.1016/s0968-0004(01)01908-9. [DOI] [PubMed] [Google Scholar]
- 58.Yu W., He X., Ni Y., Ngeow J., Eng C. Cowden syndrome-associated germline SDHD variants alter PTEN nuclear translocation through SRC-induced PTEN oxidation. Hum. Mol. Genet. 2015;24:142–153. doi: 10.1093/hmg/ddu425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 60.Scriven P., Coulson S., Haines R., Balasubramanian S., Cross S., Wyld L. Activation and clinical significance of the unfolded protein response in breast cancer. Br. J. Cancer. 2009;101:1692–1698. doi: 10.1038/sj.bjc.6605365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang G., Yang Z.Q., Zhang K. Endoplasmic reticulum stress response in cancer: molecular mechanism and therapeutic potential. Am. J. Transl. Res. 2010;2:65–74. [PMC free article] [PubMed] [Google Scholar]
- 62.Lee E., Nichols P., Spicer D., Groshen S., Yu M.C., Lee A.S. GRP78 as a novel predictor of responsiveness to chemotherapy in breast cancer. Cancer Res. 2006;66:7849–7853. doi: 10.1158/0008-5472.CAN-06-1660. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J., Jiang Y., Jia Z., Li Q., Gong W., Wang L., Wei D., Yao J., Fang S., Xie K. Association of elevated GRP78 expression with increased lymph node metastasis and poor prognosis in patients with gastric cancer. Clin. Exp. Metastasis. 2006;23:401–410. doi: 10.1007/s10585-006-9051-9. [DOI] [PubMed] [Google Scholar]
- 64.Daneshmand S., Quek M.L., Lin E., Lee C., Cote R.J., Hawes D., Cai J., Groshen S., Lieskovsky G., Skinner D.G. Glucose-regulated protein GRP78 is up-regulated in prostate cancer and correlates with recurrence and survival. Hum. Pathol. 2007;38:1547–1552. doi: 10.1016/j.humpath.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 65.Wu X.Y., Fan R.T., Yan X.H., Cui J., Xu J.L., Gu H., Gao Y.J. Endoplasmic reticulum stress protects human thyroid carcinoma cell lines against ionizing radiation-induced apoptosis. Mol. Med. Rep. 2015;11:2341–2347. doi: 10.3892/mmr.2014.2956. [DOI] [PubMed] [Google Scholar]
- 66.Khoriaty R., Vasievich M.P., Jones M., Everett L., Chase J., Tao J., Siemieniak D., Zhang B., Maillard I., Ginsburg D. Absence of a red blood cell phenotype in mice with hematopoietic deficiency of SEC23B. Mol. Cell. Biol. 2014;34:3721–3734. doi: 10.1128/MCB.00287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Korpal M., Ell B.J., Buffa F.M., Ibrahim T., Blanco M.A., Celià-Terrassa T., Mercatali L., Khan Z., Goodarzi H., Hua Y. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gellera C., Uziel G., Rimoldi M., Zeviani M., Laverda A., Carrara F., DiDonato S. Fumarase deficiency is an autosomal recessive encephalopathy affecting both the mitochondrial and the cytosolic enzymes. Neurology. 1990;40:495–499. doi: 10.1212/wnl.40.3_part_1.495. [DOI] [PubMed] [Google Scholar]
- 69.Launonen V., Vierimaa O., Kiuru M., Isola J., Roth S., Pukkala E., Sistonen P., Herva R., Aaltonen L.A. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc. Natl. Acad. Sci. USA. 2001;98:3387–3392. doi: 10.1073/pnas.051633798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bourgeron T., Chretien D., Poggi-Bach J., Doonan S., Rabier D., Letouzé P., Munnich A., Rötig A., Landrieu P., Rustin P. Mutation of the fumarase gene in two siblings with progressive encephalopathy and fumarase deficiency. J. Clin. Invest. 1994;93:2514–2518. doi: 10.1172/JCI117261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neumann H.P., Bausch B., McWhinney S.R., Bender B.U., Gimm O., Franke G., Schipper J., Klisch J., Altehoefer C., Zerres K., Freiburg-Warsaw-Columbus Pheochromocytoma Study Group Germ-line mutations in nonsyndromic pheochromocytoma. N. Engl. J. Med. 2002;346:1459–1466. doi: 10.1056/NEJMoa020152. [DOI] [PubMed] [Google Scholar]
- 72.Vanharanta S., Buchta M., McWhinney S.R., Virta S.K., Peçzkowska M., Morrison C.D., Lehtonen R., Januszewicz A., Järvinen H., Juhola M. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am. J. Hum. Genet. 2004;74:153–159. doi: 10.1086/381054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burnichon N., Brière J.J., Libé R., Vescovo L., Rivière J., Tissier F., Jouanno E., Jeunemaitre X., Bénit P., Tzagoloff A. SDHA is a tumor suppressor gene causing paraganglioma. Hum. Mol. Genet. 2010;19:3011–3020. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bourgeron T., Rustin P., Chretien D., Birch-Machin M., Bourgeois M., Viegas-Péquignot E., Munnich A., Rötig A. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 1995;11:144–149. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 75.Parfait B., Chretien D., Rötig A., Marsac C., Munnich A., Rustin P. Compound heterozygous mutations in the flavoprotein gene of the respiratory chain complex II in a patient with Leigh syndrome. Hum. Genet. 2000;106:236–243. doi: 10.1007/s004390051033. [DOI] [PubMed] [Google Scholar]
- 76.Lepoutre-Lussey C., Thibault C., Buffet A., Morin A., Badoual C., Bénit P., Rustin P., Ottolenghi C., Janin M., Castro-Vega L.J. From Nf1 to Sdhb knockout: Successes and failures in the quest for animal models of pheochromocytoma. Mol. Cell. Endocrinol. 2015 doi: 10.1016/j.mce.2015.06.027. Published online June 27, 2015. [DOI] [PubMed] [Google Scholar]
- 77.Goldenring J.R. A central role for vesicle trafficking in epithelial neoplasia: intracellular highways to carcinogenesis. Nat. Rev. Cancer. 2013;13:813–820. doi: 10.1038/nrc3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.