Abstract
Plant hormone abscisic acid (ABA) plays a crucial role in modulating plant responses to environmental stresses. Basic helix-loop-helix (bHLH) transcription factors are one of the largest transcription factor families that regulate multiple aspects of plant growth and development, as well as of plant metabolism in Arabidopsis. Several bHLH transcription factors have been shown to be involved in the regulation of ABA signaling. We report here the characterization of bHLH129, a bHLH transcription factor in Arabidopsis. We found that the expression level of bHLH129 was reduced in response to exogenously applied ABA, and elevated in the ABA biosynthesis mutant aba1-5. Florescence observation of transgenic plants expressing bHLH129-GFP showed that bHLH129 was localized in the nucleus, and transient expression of bHLH129 in protoplasts inhibited reporter gene expression. When expressed in Arabidopsis under the control of the 35S promoter, bHLH129 promoted root elongation, and the transgenic plants were less sensitivity to ABA in root elongation assays. Quantitative RT-PCR results showed that ABA response of several genes involved in ABA signaling, including ABI1, SnRK2.2, SnRK2.3 and SnRK2.6 were altered in the transgenic plants overexpressing bHLH129. Taken together, our study suggests that bHLH129 is a transcription repressor that negatively regulates ABA response in Arabidopsis.
Basic helix-loop-helix (bHLH) transcription factors are one of the largest transcription factor families find in almost all organisms, including fungi, animals and plants1. In Arabidopsis, bHLH transcription factors regulate multiple aspects of plant growth and development, as well as plant metabolism2. For example, the bHLH transcription factors GLABRA3 (GL3), ENHANCER OF GLABRA3 (EGL3) and TRANSPARENT TESTA 8 (TT8) are involved in the regulation of trichome and root hair formation, hypocotyl stomata patterning, mucilage and anthocyanin biosynthesis in Arabidopsis3,4,5,6,7, by interacting with the WD40-repeat protein TRANSPARENT TESTA GLABRA1 (TTG1) and several different R2R3 MYB proteins to form multiple MYB-bHLH-WD40 (MBW) transcription activator complexes8,9, FLOWERING BHLH (FBH) transcription activators FBH1, FBH2, FBH3 and FBH4 control flowering time by regulating the expression of the photoperiodic flowering regulator gene CONSTANS (CO)10, and PHYTOCHROME INTERACTING FACTOR 3-LIKE 5 (PIL5) regulates seed germination by activating the expression of a C3H-type zinc finger gene SOMUNS (SOM)11,12.
In addition to regulate metabolism and plant growth and development, bHLH transcription factors are also involved in the regulation of different signaling pathways including light signaling and plant hormone signaling. For example, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 (REP1), PHYTOCHROME-INTERACTING BHLH FACTORS (PIFs) and bHLH135 are involved in the regulation of light signaling13,14,15,16, MYC2 is a master regulator of jasmonate (JA) signaling and it regulates crosstalk between JA and other plant hormones17, and typical bHLH proteins including ACTIVATION-TAGGED BRI1(BRASSINOSTEROID-INSENSITIVE 1)-SUPPRESSOR 1 (ATBS1) and PACLOBUTRAZOL RESIS-TANCE1 (PRE1) are involved in the regulation of brassinosteroid signaling18,19.
Abscisic acid (ABA) is one of the earliest identified plant hormones20. Though it regulates some processes of plant growth and development such as seed germination, seed maturation and bud dormancy, ABA is largely recognized by its roles in the regulation of plant responses to environmental stresses including drought, cold, heat and salinity21,22,23. In these processes, ABA functions through a complex web of signaling networks, in which many components including receptors, phosphatases, protein kinases, E3 ligases and transcription factors are involved21,22,23,24.
Several bHLH transcription factors have been shown to be involved in ABA signaling, and the ways how bHLH transcription factors are involved in ABA signaling are different. INDUCER OF CBF EXPRESSION 2 (ICE2) induces ABA biosynthesis25, and overexpression of bHLH122 increased cellular ABA levels26. The expression of both ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR (AIB) and ANDROGEN-INDUCIBLE GENE 1 (AtAIG1) are induced by ABA27,28, however, AIB positively regulates ABA response in Arabidopsis28, whereas AtAIG1 can bind to the E-box sequence present in the promoter regions of many ABA-responsive genes, and negatively regulates ABA response27. AtMYC2 interacts with R2R3 MYB transcription factor AtMYB2 to activate the expression of ABA response gene RESPONSIVE TO DEHYDRATION 22 (RD22)29, while AtbHLH112 functions as a transcription activator and binds to GCG-box or E-box motifs to induce proline biosynthesis and ROS scavenging pathway30. On the other hand, ABA can induce the phosphorylation of three bHLH transcription factors, including ABA-RESPONSIVE KINASE SUBSTRATE 1 (AKS1), AKS2 and AKS3, leading to the repression of their transcriptional activities31.
Here we report the characterization of bHLH129, an Arabidopsis bHLH transcription factor. We showed that expression of bHLH129 was down-regulated by exogenously applied ABA. Protoplast transient transfection assay results indicated that bHLH129 is a transcription repressor. When expressed in Arabidopsis, bHLH129 promoted root elongation, and the transgenic plants overexpressing bHLH129 were less sensitive to ABA. In addition, we showed that ABA response of some ABA signaling genes including ABA INSENSITIVE 1 (ABI1), SNF1-RELATED PROTEIN KINASE 2.2 (SnRK2.2), SnRK2.3 and SnRK2.6 were altered in the transgenic plants.
Results
Expression of bHLH129 is down-regulated by ABA
Arabidopsis bHLH transcription factors have been shown to regulate plant metabolism, as well as multiple aspects of plant growth and development2. Several bHLH transcription factors are reported to regulate ABA response25,28,29,30. Our previously microarray data showed that expression of bHLH129 (At2g43140) is regulated by ABA32, suggesting that bHLH129 may involve in the regulation of ABA response.
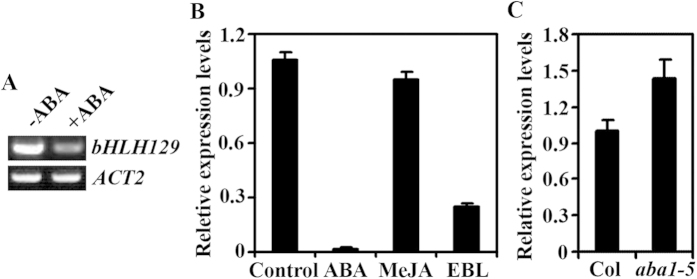
To investigate the possible roles of bHLH129 in ABA response in Arabidopsis, we examined if the expression of bHLH129 is regulated by ABA. We first examined if ABA treatment will affect the expression of bHLH129. Arabidopsis seedlings were treated with ABA for 3 h, RNA was isolated and RT-PCR or quantitative RT-PCR (qRT-PCR) was used to examine the expression of bHLH129. As shown in Fig. 1A, expression level of bHLH129 in Arabidopsis seedlings was dramatically reduced in response to exogenously applied ABA. Quantitative RT-PCR results showed that transcript levels of bHLH129 were decreased about 60-fold and 4-fold, respectively after ABA and 2,4-epibrassinolide treatments, but remained largely unchanged after methyl jasmonate treatment (Fig. 1B). We then examined if expression of bHLH129 is altered in aba1-5 (Fig. 1C), an ABA biosynthesis mutant33. We found that the transcript levels of bHLH129 increased ~1.4-fold in the aba1-5 mutant seedlings. These results suggest that bHLH129 is an ABA response gene and its expression is down-regulated by ABA.
Figure 1. Expression of bHLH129 is negatively regulated by ABA.
(A) Expression of bHLH129 in Arabidopsis seedlings in response to ABA treatment. RNA was isolated from ABA treated Arabidopsis seedlings and RT-PCR was used to examine the expression of bHLH129. Expression of ACT2 was used as a control. (B) Quantitative RT-PCR (qRT-PCR) analysis of bHLH129 expression in response to ABA, methyl jasmonate (MeJA), and 2,4-epibrassinolide (EBL) treatments. RNA was isolated from ABA, methyl jasmonate, or 2,4-epibrassinolide treated Arabidopsis seedlings, and qRT-PCR was used to examine the expression of bHLH129. Expression of ACTIN2 was used as a reference gene, and expression of bHLH129 in the absence of ABA was set as 1. Data represent the mean ± standard deviation (SD) of three replicates. (C) Expression of bHLH129 in aba1-5 mutants. RNA was isolated from 12-day-old Col and aba1-5 mutant seedlings and qRT-PCR was used to examine the expression of bHLH129. Expression of ACT2 was used as a reference gene, and expression of bHLH129 in Col wild type was set as 1. Data represent the mean ± standard deviation (SD) of three replicates.
Expression patterns of bHLH129 in Arabidopsis
To examine the expression patterns of bHLH129, different tissues and organs were collected from Arabidopsis seedlings and mature plants, and RT-PCR was used to examine the expression of bHLH129 in the samples collected. We found that transcripts of bHLH129 were detectable in all tissues and organs examined but stems, with relative higher expression levels detected in roots and cotyledons (Fig. 2A).
Figure 2. Expression pattern of bHLH129.
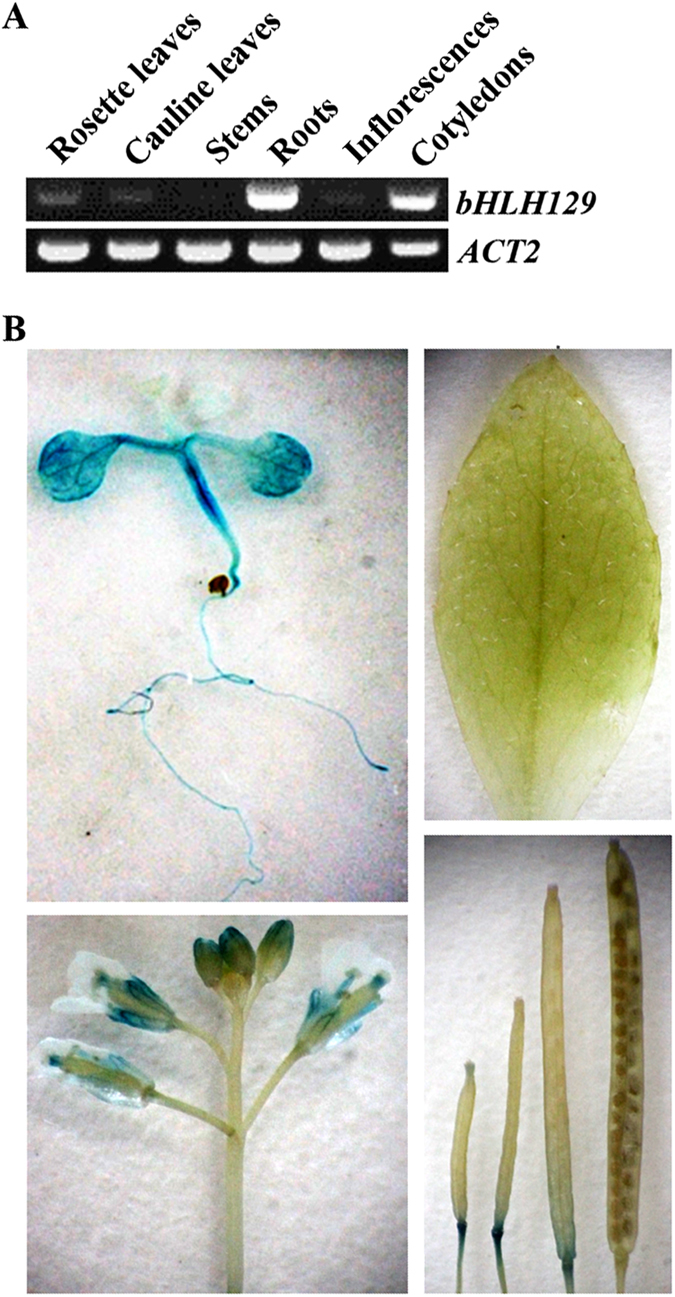
(A) Expression of bHLH129 in different tissues and organs of the Col wild type Arabidopsis. RNA was isolated from different tissues and organs of the Col wild type Arabidopsis and RT-PCR was used to examine the expression of bHLH129. Expression of ACT2 was used as a control. (B) Expression of bHLH129p:GUS in 14-day-old seedlings (upper panel, left), rosette leaves (upper panel, right), infloresences (lower panel, left) and siliques at different developmental stages (lower panel, right). X-Gluc (5-bromo-4-chloro-3-indolyl β-D-glucuronide) was used for histochemical staining of GUS activity.
To examine the expression patterns of bHLH129 in more details, we generated reporter transgenic plants by transforming wild-type plants with bHLH129p:GUS construct, and examined GUS expression in the transgenic plants. We found that, consistent with the RT-PCR results, bHLH129p:GUS was highly expressed in the roots and cotyledons, but not stems (Fig. 2B). Higher expression level of bHLH129p:GUS was also observed in hypocotyls and some flower organs (Fig. 2B). We also found that bHLH129p:GUS was expressed in young rosette leaves and the lower part of young siliques, but not mature rosette leaves and old siliques (Fig. 2B), suggesting that expression of bHLH129p:GUS appears to be developmentally regulated.
bHLH129 is a transcription repressor
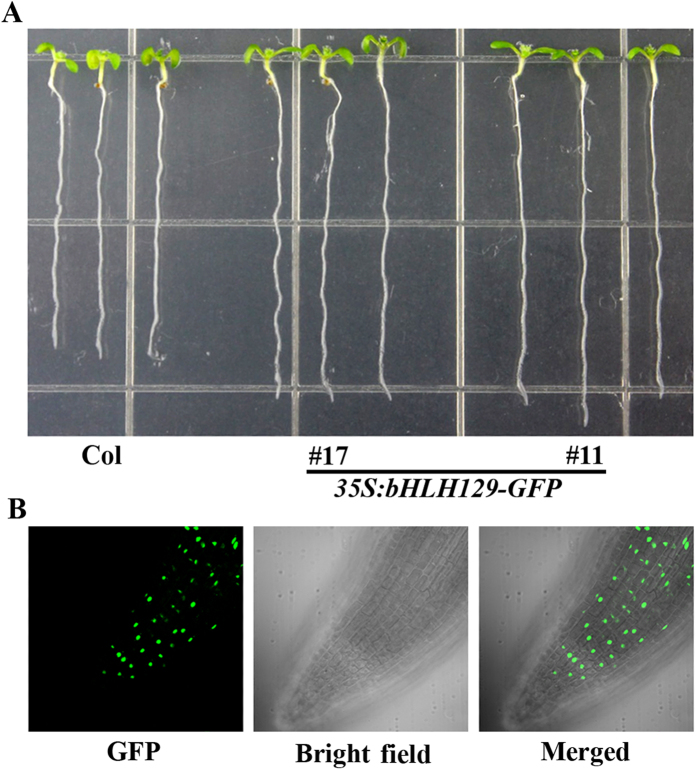
Some bHLH transcription factors such as GL3, AtbHLH112 and AtMYC2 have been shown to be transcription activators30,34, whereas some others such as JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2 and JAM3 are repressors35,36. To explore bHLH129’s functions in Arabidopsis, we decided to examine if bHLH129 functions as transcription activator or repressor. We first examined bHLH129’s subcellular localization by generating bHLH129-GFP transgenic plants and examining GFP florescence in the transgenic plants. The bHLH129-GFP transgenic plant seedlings have longer primary root when compared with that in Col wild type seedlings (Fig. 3A), a phenotype similar to that of the bHLH129 transgenic plants (see next section for details), suggesting that the bHLH129-GFP was functional. By examining GFP florescence in the root of the bHLH129-GFP transgenic plant seedlings obtained, we found that bHLH129 is predominantly localized in the nucleus (Fig. 3B).
Figure 3. Subcellular localization of bHLH129.
(A) Seven-day-old Col wild type and 35S:bHLH129-GFP transgenic plant seedlings. (B) Subcellular localization of bHLH129. Root tips of the 35S:bHLH129-GFP transgenic plant seedlings were examined under a florescence microscope. Left panel: GFP channel, middle panel: bright field image, right panel: merged.
We then examined transcriptional activities of bHLH129 using protoplast transient transfection assays. Plasmids of activator gene LD-VP, effector gene GD-bHLH129 or control gene GD, and the reporter gene LexA-Gal4:GUS were co-transfected into protoplasts, and GUS activities were measured after incubation of the transfected protoplasts overnight at darkness. In this system, the LD-VP activator can be recruited to the LexA DNA binding site thus activating the reporter gene, whereas GD control or GD-bHLH129 can be recruited to the Gal4 DNA binding site of the reporter gene. If bHLH129 functions as a transcription repressor, co-transfection of GD-bHLH129 will result in repression of the reporter gene activated by LD-VP. As shown in Fig. 4, co-transfection of the activator gene LD-VP and the control gene GD activated the reporter gene, while co-transfection of the effector gene GD-bHLH129 resulted in repression of the reporter gene, indicating that bHLH129 is a transcription repressor.
Figure 4. bHLH129 is a transcription repressor.

Effectors and reporter (diagrammed on the top of the figure) were co-transfected into protoplasts isolated from Col wild type, and the transfected protoplasts were incubated in darkness for 20-22 h before GUS activity was assayed. Data represent the mean ± SD of three replicates.
Overexpression of bHLH129 in Arabidopsis promotes root elongation
Having shown that expression of bHLH129 is down-regulated by ABA (Fig. 1), and bHLH129 functions as a transcription repressor (Fig. 4), we further explored the functions of bHLH129 by generating transgenic plants expressing bHLH129 under the control of the 35S promoter (35S:bHLH129), isolating bHLH129 knock-out mutants, and examining phenotypes of the transgenic plants and the mutants obtained.
Plant overexpressing bHLH129 was obtained by transforming Col wild type plants with the 35S:bHLH129 construct, and bHLH129 knock-out mutant bhlh129-1 was isolated from a SALK T-DNA insertion line (SALK_041780) obtained from ABRC. As shown in Fig. 5A, bHLH129 transgenic Arabidopsis seedlings have increased primary root length when compared with that of the wild type seedlings, whereas the primary root length of the bhlh129-1 mutant seedlings were largely indistinguishable from that of the wild type seedlings. Quantitative analysis showed that there was an ~20% increase in the primary root length of the transgenic plants (Fig. 5B). Overexpression of bHLH129 in the transgenic plants and knock-out of bHLH129 in the bhlh129-1 mutant were confirmed by RT-PCR (Fig. 5C).
Figure 5. Phenotypes of Arabidopsis transgenic plant seedlings expressing bHLH129.
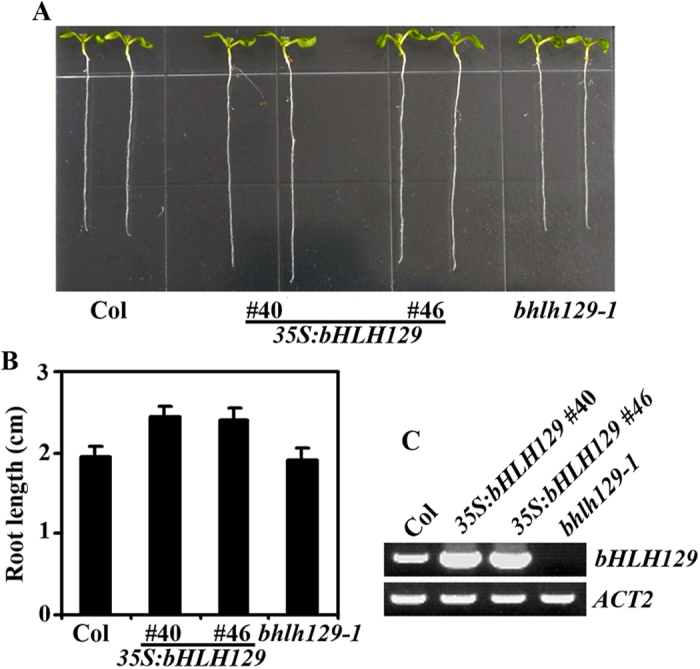
(A) Seven-day-old Col wild type and transgenic plant seedlings expressing bHLH129 under the control of the 35S promoter. (B) Primary root length of 7-day-old wild type and transgenic plants. Data represent the mean ± SD of 19-23 seedlings. (C) Expression of bHLH129 in the transgenic plants. RNA was isolated from Col wild type, bHLH129 transgenic, and bhlh129-1 mutant seedlings and RT-PCR was used to examine the expression of bHLH129. Expression of ACT2 was used as a control.
Transgenic plant seedlings expressing GFP tagged bHLH129 (35S:bHLH129-GFP) also resulted in increased primary root length (Fig. 3A), indicating that bHLH129-GFP is functional, thus the plants were used to examine subcellular localization of bHLH129 (Fig. 3B).
Transgenic plants overexpressing bHLH129 are less sensitive to ABA
Because bHLH129 is an ABA response gene (Fig. 1), we further examined if bHLH129 is involved in regulating ABA response by examining ABA responsiveness of the bHLH129 transgenic plants obtained. ABA inhibited root elongation was used to analyze ABA responsiveness in the transgenic plants. Four-day-old seedlings of wild-type and the bHLH129 transgenic plants grown on vertical 1/2 MS plates were transferred to new 1/2 MS plates at the presence and absence of ABA and grown vertically for additional 10 days. The primary root length was measured and percentage of root inhibition was calculated. As shown in Fig. 6, Arabidopsis seedlings over-expressing bHLH129 was less sensitive to ABA treatments. The root elongation of wild-type seedlings was inhibited about 50% by 5 μM ABA, whereas that of bHLH129 transgenic plants was only about 30% (Fig. 6B). Root elongation was further inhibited by 10 μM ABA, however, the bHLH129 transgenic plant seedlings were still less sensitive to ABA treatments when compared with that of the wild type seedlings (Fig. 6). On the other hand, a near wild type response to ABA treatments was observed with the bhlh129-1 mutant seedlings (Fig. 6).
Figure 6. Effects of ABA on root elongation in Col wild type and bHLH129 transgenic plant seedlings.
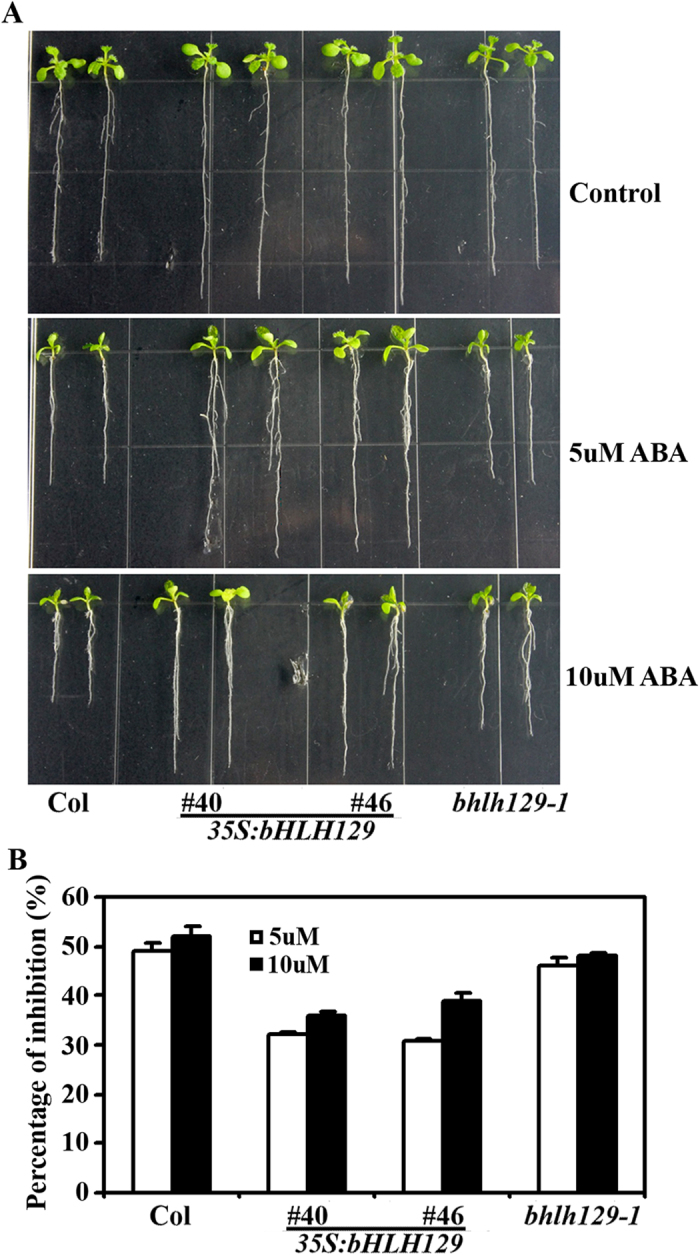
(A) Fourteen-day-old seedlings on vertical plates. Seedlings were grown vertically on 1/2 MS plates for 4 d, and then transferred to plates containing 5 μM or 10 μM ABA and grown for 10 d. (B) Root elongation inhibition by ABA. Length of new elongated roots was measured, and percentage of inhibition was calculated. Data represent means ± SD of 9-11 seedlings.
ABA response of some ABA signaling pathway genes is altered in the transgenic plants overexpressing bHLH129
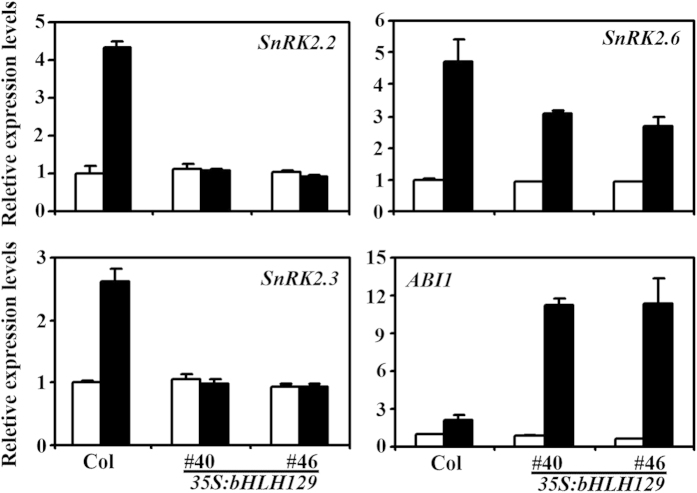
ABA signaling is regulated by several different types of proteins, including the central regulators PROTEIN PHOSPHATASE 2C (PP2C) A-group proteins and SUCROSE NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASES (SnRK2s)21,22,23,24. To investigate why the bHLH129 transgenic plants were less sensitive to ABA treatment when compared to the Col wild type, we examined the expression of some ABA signaling regulator genes including PP2C genes and SnRK2 genes in the bHLH129 transgenic plants. As shown in Fig. 7, the expression levels of SnRK2 gene SnRK2.2, SnRK2.3 and SnRK2.6, and PP2C gene ABA INSENSITIVE 1 (ABI1) in the transgenic plant seedlings were largely unaffected. When compared to that in the Col wild type plant seedlings, however, their response to ABA was altered in the transgenic plants overexpressing bHLH129 (Fig. 7). An ~2–5 folds induction of the above mentioned genes in response to ABA was observed in the Col wild type seedlings (Fig. 7). In the bHLH129 transgenic seedlings, the expression of SnRK2.2 and SnRK2.3 was no longer induced by ABA treatment. On the other hand, expression levels of SnRK2.6 in response to ABA in the transgenic plant seedlings was reduced to about two-third of that in the wild type seedlings, whereas the expression levels of ABI1 was increased about 4 folds (Fig. 7).
Figure 7. Expression of the genes involved in ABA signaling in wild type and bHLH129 transgenic plant seedlings.
RNA was isolated from ABA treated Col wild type and bHLH129 transgenic plant seedlings, and qRT-PCR was used to examine the expression of the genes involved in ABA signaling. Expression of ACT2 was used as a reference gene, and expression of corresponding genes in Col wild type seedlings in the absence of ABA was set as 1. Data represent the mean ± SD of three replicates. White bar, absence of ABA; black bar, presence of ABA.
Discussion
There are 162 genes in Arabidopsis encoding bHLH transcription factors37. So far only a few of them have been reported to be involved in the regulation of ABA response25,28,29,30. Our results here show that bHLH129 is an ABA response gene, it encodes a transcription repressor, and is involved in the regulation of ABA response in Arabidopsis.
Among the several bHLH transcription factor genes that involve in the regulation of ABA response in Arabidopsis, AIB and AtAIG1 have been shown to be up-regulated by ABA27,28, overexpression of bHLH122 increases cellular ABA levels, but bHLH122 itself is not induced by ABA26. Two different pieces of evidence suggest that bHLH129 is an ABA response gene, but its expression is down-, rather than up-regulated by ABA: expression levels of bHLH129 were reduced in the presence of exogenously applied ABA, and elevated in the ABA biosynthesis mutant aba1-5 (Fig. 1).
Several bHLH transcription factors that regulate ABA response in Arabidopsis including bHLH112 and AtMYC2 have been shown to be transcription activator29,30. AIB/JIM1 is initially showed as a transcription activator in yeast cells28, but assays in plant cells suggest that AIB/JIM1 and its homologues JAM2 and JAM3 function as transcription repressor, and they are involved in the regulation of JA signaling35,36. We found that bHLH129 is a nucleus protein (Fig. 3), when transient expressed in protoplasts, it repressed the expression of the reporter gene activated by a transcription activator, indicating that bHLH129 is a transcription repressor.
When overexpressed in Col wild type plants, bHLH129 promoted root elongation (Fig. 5), and the bHLH129 transgenic plants were less sensitive to ABA in root elongation assays (Fig. 6), suggesting that bHLH129 is a negative regulator of ABA response. Considering that ABA down-regulate the expression of bHLH129, while bHLH129 negatively regulates ABA response, it is likely that bHLH129 plays a role under normal growth conditions when ABA response should not be triggered. In Col wild type plants, application of ABA resulted in decreased expression of bHLH129, the plants showed a normal ABA response. In the 35S:bHLH129 transgenic plants, expression of bHLH129 is no longer down-regulated by ABA, thus the plants showed a reduced response to ABA treatment. However, bHLH129 knock-out mutant bhlh129-1 is morphological similar to Col wild type plants (Fig. 5), and it showed a near wild type response to ABA. This may due to redundancy functions of other related bHLH transcription factors.
The mechanisms bHLH transcription factors used to regulate ABA signaling are different, some of them including ICE2 and bHLH122 affect ABA biosynthesis25,26, and some others such as AtAIG1 and AtMYC2 directly regulate the expression of ABA response genes27,29.
PP2Cs and SnRK2s are key central regulators in ABA signaling pathway21,22,23,24. We found that although the expression levels of PP2Cs and SnRK2s genes including ABI1, SnRK2.2, SnRK2.3 and SnRK2.6 in the transgenic plants overexpressing bHLH129 were largely unaffected when compared with that in the Col wild type plants at the absence of ABA, however, their response to ABA was altered (Fig. 7), indicating that bHLH129 may regulate ABA response through regulating the expression of some ABA signaling pathway genes. Because bHLH129 function as a transcription repressor (Fig. 4), and ABA response of SnRK2s genes in the transgenic plants was repressed, while that of PP2C gene ABI1 was enhanced (Fig. 7), it is very unlikely that all these genes were sonly regulated by bHLH129. Considering that bHLH transcription factors have been reported to interact with other transcription factors to regulate several different processes including cell fate determination, mucilage and anthocyanin biosynthesis, and to regulate the expression of ABA response gene RD228,9,29, bHLH129 may interact with other transcription factors to regulate ABA response in Arabidopsis. In any case, it will be of great interest to find out how bHLH129 regulates the expression of those genes, and why the effects of bHLH129 on the expression of ABA signaling pathway genes can only be seen in the presence of ABA.
Never the less, our results showed that expression of bHLH129 is down-regulated by ABA, bHLH129 is a transcription repressor, and it regulated root elongation and ABA response when overexpressed in Arabidopsis, possibly by regulating the expression of some ABA signaling pathway genes.
Methods
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used for plant transformation and protoplasts isolation. The bhlh129-1 mutant was isolated from a SALK T-DNA insertion line (SALK_041780) obtained from ABRC.
To obtain seedlings for phenotypic analysis and ABA treatment, Arabidopsis seeds were sterilized and sown on ½ MS (Murashige & Skoog) plates with vitamins (PlantMedia) and 1% (w/v) sucrose, solidified with 0.6% (w/v) phytoagar (PlantMedia). The plates were kept at 4 °C in darkness for 2 days before moved into a growth room. To obtain plants for plant transformation and protoplasts isolation, Arabidopsis seeds were sown directly into soil pots and kept in a growth room. The growth condition in the growth room was set with a temperature at 22 °C and a photoperiod with 16 h/8 h (light/dark) at a light density of approximately 120 μmol m−2s−1.
ABA treatment and root elongation assay
To examine the expression of bHLH129 and genes involved in ABA signaling in response to ABA, 12-day-old Col wild type seedlings were treated with 10 μM ABA, 200 nM methyl jasmonate, or 100 nM 2,4-epibrassinolide for 3 h in darkness on a shaker at 40 rpm. Samples were frozen in liquid N2 and kept at −80 °C for RNA isolation.
To examine ABA sensitivity of the bHLH129 transgenic plant seedlings, 4-day-old Col wild type and bHLH129 transgenic plant seedlings grown on vertical plates were transferred into new plates containing either 5 μM or 10 μM ABA and grown vertically. Pictures were taken 10 days after transfer, root length was measured using Image J software, and percentage of inhibition was calculated. The experiments were repeated three times, and data from a representative experiment was presented.
DNA and RNA isolation, RT-PCR and quantitative RT-PCR (qRT-PCR)
DNA and total RNA from Arabidopsis seedlings were isolated as described previously9,38,39,40. cDNA was synthesized using 2 μg total RNA by Oligo(dT)-primed reverse transcription using the EazyScript First-Strand DNA Synthesis Super Mix (TransGen Biotech).
RT-PCR was used to examine the expression of bHLH129, qRT-PCR was used to examine the expression of bHLH129 and genes involved in ABA signaling pathway. Arabidopsis gene ACTIN2 (ACT2) was used as a control for RT-PCR and qRT-PCR. Primers used for RT-PCR and qRT-PCR analysis of bHLH129 and genes involved in ABA signaling pathway are: bHLH129, 5′-TTTCTCTAGGACGGCCAAAC-3′ and 5′-GATGGCTACTACTCCCACTAGA-3′, SnRK2.2, 5′-CCGATTATGCACGACAGTGA-3′ and 5′-CAAGCTCCTTGGTGACTCTATC-3′, SnRK2.3, 5′-GAAGATCCAGAAGAGCCAAGAG-3′ and 5′-CCGTATGTCATCAGGGATTGAG-3′, SnRK2.6, 5′-CGGGCCAAAGCATAGAAGAA-3′ and 5′-CCAAGCTTCCTGTGAGGTAATG-3′. The primers for qRT-PCR examination of ABI1, and for RT-PCR and qRT-PCR examination of ACT2 have been described previously9,40,41.
Constructs
The reporter construct LexA-Gal4:GUS, and effector constructs GD and LD-VP used for protoplast transfection have been described previously42,43.
To generate GD tagged bHLH129 construct for protoplast transfection assays and HA tagged bHLH129 construct for plant transformation, the full-length open-reading frame (ORF) of bHLH129 was amplified by RT-PCR using RNA isolated from the Arabidopsis seedlings, and cloned in frame with an N-terminal GD or HA tag into the pUC19 vector under the control of the 35S promoter44,45. HA tagged bHLH129 construct in pUC19 was then digested with proper enzymes, and subcloned into the binary vector pPZP21146.
To generate GFP tagged construct for subcellular localization analysis of bHLH129, the ORF of bHLH129 was amplified and cloned in frame with a C-terminal GFP tag into the pUC19, the construct obtained was then digested with proper enzymes, and subcloned into the binary vector pPZP211 for plant transformation.
To generate bHLH129p:GUS (β-glucuronidase) construct for plant transformation, a 2514 bp DNA fragment immediately before the start condon of bHLH129 was amplified by PCR using DNA isolated from Arabidopsis seedlings, and used to replace the OFP1 promoter in the pPZP211OFP1p:GUS construct43.
The primers used to generate GD/HA-bHLH129 construct are 5′-CAACATATGTACCCTCCTAATTCCTC-3′ and 5′-CAAGAGCTCTCATCGCTTCTTGCATGC-3′, the primers used to generate bHLH129-GFP construct are 5′-CAACATATGTACCCTCCTAATTCCTC-3′ and 5′-CAAGAGCTCTCGCTTCTTGCATGC-3′, and the primers used for make bHLH129p:GUS construct are 5′-CAACTGCAGCATGATCGGTCTGATTC-3′ and 5′-CAAGAGCTCGAAACCGGAAAAGAAAAACCC-3′.
Plant transformation and transgenic plants selection
About 5-week-old plants with several mature flowers on the main inflorescence were used for transformation by using the floral dip method47. Transgenic plants were selected by grown T1 seeds on 1/2 MS plates containing 50 μg/ml kanamycin and 100 μg/ml carbenicillin. For each construct, at least 5 transgenic lines with similar phenotypes were obtained, and represent homozygous T3 or T4 plants were used for further analysis.
Plasmid DNA isolation, protoplasts isolation, transfection and GUS activity assays
Plasmids preparation, protoplasts isolation, transfection and GUS activity assay were performed as described previously9,39,40,43,44,45,48. In brief, reporter and effector plasmid DNA were isolated using the GoldHi EndoFree Plasmid Maxi Kit (CWbiotech), and co-transfected into protoplasts isolated from rosette leaves collected from ~4-week-old wild type Arabidopsis plants. Transfected protoplasts were incubated under darkness at room temperature for 20–22 h, and then GUS activities were measured by using a SynergyTM HT microplate reader (BioTEK).
GUS staining
GUS activity was monitored by staining seedlings and different organs of adult bHLH129p:GUS transgenic Arabidopsis plants with X-gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronide, Rose Scientific Ltd.) using the procedure described previously49.
Microscopy
Photographs of Col wild type and transgenic Arabidopsis seedlings were taken under a Motic K dissection microscope equipped with an EOS 1100D camera. GFP florescence of bHLH129-GFP transgenic seedlings was examined, and photographs were taken under an Olympus FV1000 confocal microscope.
Additional Information
How to cite this article: Tian, H. et al. An ABA down-regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis. Sci. Rep. 5, 17587; doi: 10.1038/srep17587 (2015).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31470297), the Key Laboratory of Molecular Epigenetics of MOE (130014542), and the Programme for Introducing Talents to Universities (B07017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions S.W. conceived the study. H.T. and S.W. designed the experiments. H.T., H.G., X.D., Y.C., K.Z. and X.W. performed the experiments. H.T., H.G. and S.W analyzed the data. S.W. drafted the manuscript, and all authors read and approved the final manuscript.
References
- Skinner M. K., Rawls A., Wilson-Rawls J. & Roalson E. H. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation. 80, 1–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Li X. & Ma L. Basic helix-loop-helix transcription factors and epidermal cell fate determination in Arabidopsis. Plant Signal Behav. 7, 1556–1560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N. et al. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 12, 1863–1878 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C. T., Zhang F. & Lloyd A. M. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics. 156, 1349–1362 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C. et al. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development. 130, 6431–6439 (2003). [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Zhao M., Gonzalez A., Lloyd A. & Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 132, 291–298 (2005). [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J. & Torii K. U. Breaking the silence: three bHLH proteins direct cell-fate decisions during stomatal development. Bioessays. 29, 861–870 (2007). [DOI] [PubMed] [Google Scholar]
- Lin Q. & Aoyama T. Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: update on the roles of the classic regulator. J Integr Plant Biol. 54, 729–737 (2012). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 83, 300–311 (2015). [DOI] [PubMed] [Google Scholar]
- Ito S. et al. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl Acad. Sci. USA 109, 3582–3587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E. et al. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 16, 3045–3058 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee N., Kim W., Lim S. & Choi G. ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell. 23, 1404–1415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh M. S., Kim Y. M., Han S. J. & Song P. S. REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in arabidopsis. Plant Cell. 12, 2061–2074 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday K. J., Hudson M., Ni M., Qin M. & Quail P. H. poc1: an Arabidopsis mutant perturbed in phytochrome signaling because of a T-DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc. Natl Acad. Sci. USA. 96, 5832–5837 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E. & Quail P. H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21, 2441–2450 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P. et al. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell. 24, 1398–1419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. & Manners J. M. MYC2: the master in action. Mol Plant. 6, 686–703 (2013). [DOI] [PubMed] [Google Scholar]
- Wang H. et al. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell. 21, 3781–3791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Y. et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 21, 3767–3780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies P. J. Plant Hormones: Physiology, Biochemistry and Molecular Biology. Dordrecht: Kluwer (1995). [Google Scholar]
- Shang Y. et al. The Mg-chelatase H subunit of arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 22, 1909–1935 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T. et al. Molecular basis of the core regulatory network in aba responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton D. L. et al. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J. 10, 2–11 (2012). [DOI] [PubMed] [Google Scholar]
- Guo J., Yang X., Weston D. J. & Chen J. G. Abscisic acid receptors: past, present and future. J Integr Plant Biol. 53, 469–479 (2011). [DOI] [PubMed] [Google Scholar]
- Kurbidaeva A., Ezhova T. & Novokreshchenova M. Arabidopsis thaliana ICE2 gene: phylogeny, structural evolution and functional diversification from ICE1. Plant Sci. 229, 10–22 (2014). [DOI] [PubMed] [Google Scholar]
- Liu W. et al. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 201, 1192–1204 (2014). [DOI] [PubMed] [Google Scholar]
- Kim J. & Kim H. Y. Molecular characterization of a bHLH transcription factor involved in Arabidopsis abscisic acid-mediated response. Biochim Biophys Acta. 1759, 191–194 (2006). [DOI] [PubMed] [Google Scholar]
- Li H. et al. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol Biol. 65, 655–665 (2007). [DOI] [PubMed] [Google Scholar]
- Abe H. et al. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 15, 63–78 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 207, 692–709 (2015). [DOI] [PubMed] [Google Scholar]
- Takahashi Y. et al. bHLH transcription factors that facilitate K. uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci Signal. 6, ra48 (2013). [DOI] [PubMed] [Google Scholar]
- Guo J. et al. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 155, 370–383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J. & Mauch-Mani B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38, 119–130 (2004). [DOI] [PubMed] [Google Scholar]
- Wang S. & Chen J. G. Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol. 49, 1792–1804 (2008). [DOI] [PubMed] [Google Scholar]
- Nakata M. & Ohme-Takagi M. Two bHLH-type transcription factors, JA-ASSOCIATED MYC2-LIKE2 and JAM3, are transcriptional repressors and affect male fertility. Plant Signal Behav. 8, e26473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M. et al. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell. 25, 1641–1656 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E. L. & Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116 (2011). [DOI] [PubMed] [Google Scholar]
- Wang S. et al. Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis. Sci Rep. 4, 5054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H. et al. An auxin responsive CLE gene regulates shoot apical meristem development in Arabidopsis. Front Plant Sci. 6, 295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. et al. Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front Plant Sci. 6, 388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y. et al. The tumor necrosis factor receptor-associated factor (TRAF)-like family protein SEVEN IN ABSENTIA 2 (SINA2) promotes drought tolerance in an ABA-dependent manner in Arabidopsis. New Phytol. 202, 174–187 (2014). [DOI] [PubMed] [Google Scholar]
- Tiwari S. B., Hagen G. & Guilfoyle T. J. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell. 16, 533–543 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chang Y., Guo J. & Chen J. G. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 50, 858–872 (2007). [DOI] [PubMed] [Google Scholar]
- Tiwari S. B., Hagen G. & Guilfoyle T. J. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 15, 533–543 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tiwari S. B., Hagen G. & Guilfoyle T. J. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell. 17, 1979–1993 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z. & Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 25, 989–994 (1994). [DOI] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- Zhou L. et al. Control of trichome formation in Arabidopsis by poplar single-repeat R3 MYB transcription factors. Front Plant Sci. 5, 262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G. & Guilfoyle T. J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 9, 1963–1971 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]