Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest shell.
| Sa_enolase | Sa_enolase–PEP | |
|---|---|---|
| Data collection | ||
| Space group | P4212 | I4 |
| Unit-cell parameters | ||
| a = b (Å) | 164.7 | 145.15 |
| c (Å) | 77.3 | 100.51 |
| α = β = γ (°) | 90 | 90 |
| Resolution range (Å) | 50–2.45 (2.54–2.45) | 50–1.60 (1.63–1.60) |
| No. of unique reflections | 39128 | 136548 |
| Wilson plot B factor (Å2) | 38.5 | 11.2 |
| R meas † (%) | 8.6 (51.9) | 9.2 (49.8) |
| Mean I/σ(I) | 29.2 (4.5) | 40.3 (6.8) |
| Completeness (%) | 98.3 (99.4) | 100.0 (100.0) |
| Multiplicity | 7.3 (7.4) | 8.2 (8.1) |
| Refinement | ||
| Resolution range (Å) | 50–2.45 | 50–1.60 |
| R work ‡/R free § (%) | 18.55/22.86 | 14.84/16.35 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.009 |
| Bond angles (°) | 1.286 | 1.376 |
| B factors (Å2) | ||
| Protein | 45.97 | 15.46 |
| Water | 36.01 | 26.73 |
| Mg2+ | 33.74 | 11.12 |
| Other ligands | 59.36 | 29.52 |
| Ramachandran plot | ||
| Most favoured regions (%) | 97.5 | 97.8 |
| Additionally allowed regions (%) | 2.3 | 2.0 |
| Outliers (%) | 0.2 | 0.2 |
R
meas was estimated by multiplying the conventional R
merge value by the factor [N/(N − 1)]1/2, where N is the data multiplicity; R
merge = 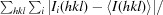
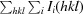 , where Ii(hkl) is the intensity of the ith measurement and 〈I(hkl)〉 is the mean intensity for that reflection.
, where Ii(hkl) is the intensity of the ith measurement and 〈I(hkl)〉 is the mean intensity for that reflection.
R = 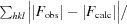
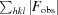 , where |F
obs| and |F
calc| are the observed and calculated structure-factor amplitudes, respectively.
, where |F
obs| and |F
calc| are the observed and calculated structure-factor amplitudes, respectively.
R free was calculated with 5.0% of the reflections in the test set.
