Abstract
Quantitative magnetic resonance imaging (MRI) techniques have the ability to quantitatively report various pathophysiological processes associated with cancer. These measures have been shown to provide complementary information to that typically obtained from standard morphologically based criteria (e.g., size) and, furthermore, have been shown to outperform sized based measures in certain applications. In this review, we discuss eight areas of quantitative MRI that are either currently employed in clinical trials, or are emerging as promising techniques for both diagnosing cancer as well as assessing—or even predicting—the response of cancer to various therapies. The currently employed methods include the response evaluation criteria in solid tumors (RECIST), dynamic susceptibility MRI (DSC-MRI), dynamic contrast enhanced MRI (DCE-MRI), and diffusion weighted imaging (DWI). The emerging techniques covered are chemical exchange saturation transfer MRI (CEST-MRI), elastography, hyperpolarized MRI, and multi-parameter MRI. After a brief introduction to each technique, we present a small number of illustrative applications before noting the existing limitations of each method and what must be done to move each to more routine clinical application.
Keywords: DCE-MRI, DSC-MRI, diffusion, CEST, elastography, hyperpolarized, multi-parametric
1. Incorporating Quantitative MRI in clinical trials
The last decade has seen tremendous interest and research effort devoted to the use of quantitative imaging within oncology (1,2). Quantitative imaging techniques can measure various properties within medical images that might serve as reliable surrogates for various pathophysiological processes with which to personalize cancer therapy and accelerate drug development (3). Furthermore, the prospect of combining several quantitative imaging measures for establishing radiologic phenotypes predictive of clinical trajectories is particularly appealing, and magnetic resonance imaging (MRI) has emerged as a promising modality for this purpose (4). MRI continues to be a mainstay for conventional size-based tumor assessments (i.e., the Response Evaluation Criteria in Solid Tumors, RECIST; see next section) that are standard efficacy endpoints within clinical trials and increasingly common in routine, standard-of-care settings. Moreover, MRI applications that can quantitatively report on various aspects of tumor biology, including perfusion, cellularity, metabolism, and protein deposition, offer the potential to supplement and enhance conventional anatomic information, which when used alone provides an incomplete assessment of solid tumors (5). This review spotlights some of the leading technological developments in MR that are laying the groundwork for quantitative MRI to transition from being viewed as an advanced research paradigm to becoming a widely established clinical reality for the cancer community. This transition presents a number unique challenges as well as exciting opportunities for imaging science.
There are several key steps for the development and evaluation of a particular quantitative imaging measure before it can be considered a true “biomarker” and safely incorporated into clinical practice (6; please see Box 1). Perhaps one of the most essential tools for the evaluation of biomarkers is the multicenter clinical trial. Over the last several years the National Cancer Institute (NCI) has spearheaded efforts to coordinate multicenter biomarker studies for imaging. The NCI’s Quantitative Imaging Network currently includes 17 Centers of Imaging Excellence in the US, several of which are actively engaged in validation of imaging based biomarkers (9). Other groups, including the Radiological Society of North America Quantitative Imaging Biomarker Alliance (RSNA QIBA) (1) and the American College of Radiology Imaging Network, recently merged with the Eastern Cooperative Oncology Group to form ECOG-ACRIN (10), have also been rigorously pursuing multicenter quantitative imaging clinical trials. These groups have developed their own clinical trial designs and workflows, image acquisition and analysis procedures, and regulatory processes. There are also efforts to harmonize procedures and practices across these groups in order to arrive at a comprehensive set of standards for the clinical validation and implementation of quantitative imaging biomarkers. We draw from their collective insights to emphasize a few key commonalities with our own experiences and discuss some administrative, regulatory, and logistical considerations facing trials of a putative MRI biomarker.
Box 1. Key steps for the development and evaluation of quantitative imaging measures.
Validation tests the accuracy, precision, repeatability, and reproducibility of the biomarker measurement
Qualification establishes the biomarker as a surrogate for tumor pathophysiology, response to therapy, or other clinical endpoint of interest
Utilization examines the performance and implementation of the biomarker within the specific context of its proposed use, especially across multiple institutions and clinical settings.
One of the most crucial aspects of successful integration of MRI biomarker research with an oncology clinical trial will be the level of engagement and collaboration the imaging scientists and radiologists have with the medical oncologists and clinical trial sponsors. Investigator-initiated trials offer certain advantages in this regard relative to industry-sponsored trials, since the latter often require a higher level of engagement and a greater emphasis on allocating resources for data management and regulatory compliance. Furthermore, since these studies are frequently designed at the industrial sponsor months—or years—before academic investigators become aware of it, integrating an advanced imaging technique can be difficult. Regardless of the type of trial, there are several characteristics pertaining to the design and execution of an MR-based imaging biomarker study that need to be considered within the context of a therapeutic oncology trial. Ideally, the biomarker study design would be rationally tailored to address a clearly defined clinical problem (e.g., predicting which patients will benefit from neoadjuvant therapy) and would test the ability of a candidate MRI technique, or group of techniques, to predict or correlate with a desired clinical outcome (e.g., pathologic complete response). The predictive value of a particular imaging measure will likely vary depending on the choice of clinical endpoint. When progression-free or overall survival is a primary endpoint, incorporation of multiple strategically chosen imaging time points during follow-up is recommended (please see Box 2).
Box 2. Helpful factors when determining the timing of follow-up scans.
The expected mechanism, onset, and duration of action of the therapeutic agent or intervention under investigation in the clinical trial.
The schedule of events (i.e., timing of biomarker scan should coincide whenever possible with the patient’s clinical appointments) within the trial.
Potential interference from use of contrast media in other clinical trial radiological procedures.
Interscan interval in relation to reimbursement policies of the sponsor, patient’s insurance provider, and/or Centers for Medicate and Medicaid Services (CMS).
The first scientific body to review, advise, and ultimately approve an imaging based biomarker study is the institutions’ Scientific Review Committee (SRC), which consists of clinicians, basic scientists, biostatisticians, nurses, pharmacists, and other medical professionals whose primary mission is to scrutinize the scientific merit and clinical prioritization of a new study in the context of an institution’s existing menu of studies. For most institutions a new clinical trial protocol will undergo review by the SRC before it is reviewed by the local Institutional Review Board (IRB). A critical aspect of the SRC review entails thorough scrutiny of the statistical methodology proposed in the study. Prior to protocol submission it is highly worthwhile, and at some institutions required, to meet with a qualified biostatistician, particularly one well versed in the analysis of imaging data, to ensure that the aims and study design are in keeping with a sound statistical framework appropriate for the development of imaging biomarkers (15). Studies must be demonstrated as having accrual goals capable of satisfying a predefined level of statistical power (typically, 80%) to determine the predictive association between the MR biomarker(s) in question and the primary clinical endpoint. Sample sizes must also be justified on the basis of historical accrual data within identical or very similar patient populations and clinical settings. Feedback from the SRC is one of the primary opportunities for constructive criticism so that the aims, design, and future conduct of a study is consistent with institutional standards for statistical rigor, clinical relevance, and scientific quality.
Once SRC approval is obtained, the next step in opening a new imaging study is protocol review and approval by the local IRB. Whereas scientific rigor and clinical relevance are the primary concerns in SRC review, the IRB is typically most focused on ensuring that the study meets all federal, state, and local policies pertaining to patient safety and confidentiality. For MRI studies one of the most important aspects of IRB review will be centered on the patient screening process to ensure compatibility with the large magnetic field the patient will encounter as part of the imaging procedure. The IRB will verify that prospective patients will be given every means necessary to disclose the presence of ferromagnetic materials, and will also allow designated key study personnel to access the prospective patient’s medical record to verify MR compatibility of implants. This is especially important in cancer patients receiving chemotherapy and/or other procedures (biopsy, surgery), since vascular access ports, biopsy marker clips, and stents are constructed of materials whose MR compatibility can vary widely. The IRB will heavily scrutinize methods for verifying the manufacturer and model number for an implant or device in question. Rigorous rating standards from the American Society for Testing and Materials (ASTM) International exist for virtually all implantable biomaterials and have been approved by the FDA. The IRB will mandate that only implanted materials having an ASTM rating of “MR Safe” at the particular field strength in question are included within the study. Implants having a rating of “MR Conditional” are often excluded, but there are instances where a particular MRI environment with specific conditions may be acceptable. For example, at 3.0T, the only implants or devices currently accepted on study are those classified as MR Conditional 6 (ASTM Standard F2503) (16).
While the IRB is reviewing the imaging protocol, there are several steps that can be taken to help ensure no delays are experienced in opening to accrual. Like therapeutic trials, studies devoted to validating a perspective imaging biomarker should be nationally registered at ClinicalTrials.gov and/or the NCI. At many institutions, national study registration is required of all clinical studies as a matter of local policy, and recent changes to the registration rules put forward by the International Committee of Medical Journal Editors now necessitate registration even for non-interventional imaging studies. Another critical logistical step prior to opening to accrual is to ensure that all imaging-related study materials have been distributed to the appropriate study personnel. The most convenient venue for such interactions is the Site Initiation Visit, where imaging scientists, clinicians, research nurses and others can meet to review key aspects of the study to establish an adequate recruitment plan and clinical workflow. This is particularly important in situations where the acquisition of imaging data is to occur on dedicated research scanners in facilities that are separated from the cancer clinics where recruitment will take place, since the processes and procedures involved in advanced imaging studies are often unfamiliar to clinical staff.
2. Current Use of MRI for Clinical Trials
In modern clinical oncology practice, MRI is widely used as a tool for cancer screening, lesion detection, lesion characterization, and therapy monitoring. Within cancer clinical trials MRI is employed for assessing response to treatment, although its role varies depending on the anatomical site of disease. For many clinical trials in solid malignancies, MRI may play a secondary role to computed tomography (CT) and may be used only when there is a contraindication to iodinated intravenous contrast media. For certain tumor types (e.g., brain and head/neck cancers), MRI may be the preferred modality for response assessment due to its excellent soft tissue contrast resolution.
When MRI is used for treatment response assessment, most current cancer clinical trials use one of several standardized response assessment guidelines based on changes in gross lesion size. These guidelines specify how to identify and measure target lesions at baseline imaging prior to therapy, how to evaluate disease burden at follow-up time points following initiation of treatment, and how to place patients into response categories at successive time points over the course of the clinical trial (17). The most widely used response assessment guideline, incorporated into most modern solid tumor clinical trials, is the Response Evaluation Criteria in Solid Tumors (RECIST) (18). As its emphasis is on changes in tumor size measurement over time, RECIST necessitates high-spatial resolution MR imaging techniques optimized for capturing anatomical detail.
Measuring changes in tumor size on anatomical imaging has been the mainstay of imaging-based response assessment for decades (19) and is supported by research linking tumor shrinkage in early-stage trials with subsequent survival benefits (20–23). However, an exclusive focus on anatomical imaging has recently been called into question with the emergence of functional imaging techniques that provide information on tumor status beyond lesion size. These techniques, many of which are MR-based and are described below, offer the promise of reporting on response at an earlier time point than traditional tumor size-based approaches, which may lag weeks to months behind a physiologic tumor response. Functional imaging techniques may also succeed in better capturing and measuring the antitumor efficacy of newer targeted agents, the cytostatic effects of which may be underestimated by traditional size-based approaches.
3. Quantitative MRI Techniques Currently Available for Clinical Trials
Here we focus on three techniques that have advanced to the point where they are frequently used in clinical trials to report on therapeutic response. In section 4, we focus on four emerging techniques.
3.1 Dynamic susceptibility contrast MRI
Abnormal angiogenesis is a common characteristic of malignant brain tumors and dynamic susceptibility contrast MRI (DSC-MRI) is frequently employed to non-invasively interrogate the hemodynamic features of the expanding vascular network. In DSC-MRI, dynamic MR images are acquired before and after an intravenous bolus injection of a contrast agent (CA), which is typically one of several clinically, approved gadolinium chelates. As the CA passes through tissue it decreases the relaxation times (T1, T2, and T2*) of tissue water and the associated MRI signal intensity. The magnitude of the change in the relaxation rate is determined by the concentration of the CA and the geometry of the tissue structures containing the CA. Pharmacokinetic models can be applied to DSC-MRI data to estimate blood volume (BV), blood flow (BF), and mean transit time (MTT) (24–26).
Given the known association between brain tumor pathology and angiogenesis, early DSC-MRI studies demonstrated the clinical utility of this technique by verifying a positive correlation between tumor blood volume and brain tumor grade (25, 27–31). As an example, Boxerman et al found relative blood volume values (i.e., relative to normal appearing white matter) of 1.52, 2.84, and 3.96 in a cohort of patients with WHO grades II (n = 11), III (n = 9) and IV (n = 23), respectively (29). Further, the correlation between blood volume and tumor grade was significant (r = 0.60; P < 0.0001). It was noted that designating tumor grade based on blood volume maps alone, however, may be confounded by intragrade variability, particularly between grades III and IV.
While such diagnostic studies served to support the consideration of DSC-MRI in brain tumor patient management, its clinical potential was more fully realized when studies emerged demonstrating its prognostic capabilities. Law et al investigated the ability of pre-treatment blood volume maps to predict clinical response (complete response, stable disease, progressive disease and death) in patients with low-grade gliomas undergoing standard-of-care treatments (32). The patients with lesions exhibiting blood volume values less than 1.75 (relative to normal appearing white matter) had a median time to progression of 4620 days, whereas those with values higher than 1.75 had a median time to progression of 245 days. An important conclusion in this study is that while DSC-MRI may have low specificity for diagnosing low-grade gliomas, it has a much higher specificity for predicting clinical endpoints in patients receiving standard treatment regimes.
In the context of routine therapy and clinical trials, standard MRI techniques are unable to reliably differentiate between progressive disease (PD) and pseudoprogression (PsP). Due to the heightened angiogenic response in recurring gliomas, DSC-MRI derived CBV maps have been explored as a means to overcome this limitation (33–35). In a Phase II clinical trial of temozolomide, paclitaxel poliglumex, and concurrent radiation, the mean CBV measured at initial progressive enhancement and the change in CBV after therapy were used to distinguish PD and PsP (33). The single time point CBV values acquired after therapy were similar between patients exhibiting PsP and PD (2.35 vs. 2.17, p = 0.67). However, changes in CBV between follow-up examinations were significantly different between PsS and PD (−0.84 and 0.84, p = 0.001) as were the trends in CBV (negative vs. positive slope; p = 0.04). It was concluded that longitudinal changes in post-therapy CBV values may be more useful for tracking treatment response as compared to static values (figure 1).
Figure 1.
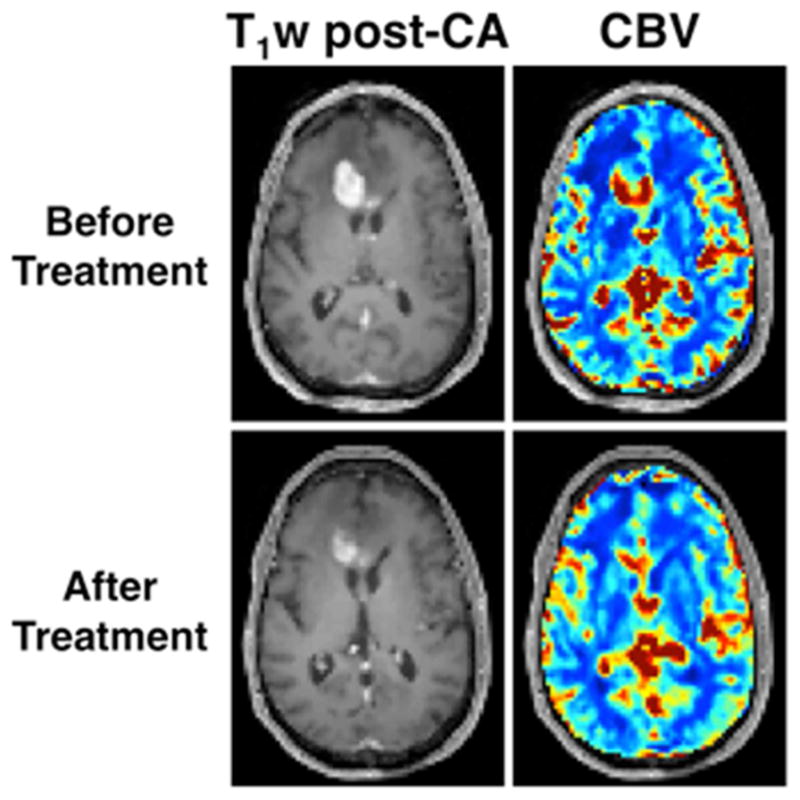
Example DSC-MRI based assessment of Bevacizumab induced cerebral blood volume (CBV) changes in recurrent high grade glioma. Two weeks of Bevacizumab treatment reduced contrast agent extravasation and the enhancing tumor volume (left column). Treatment also decreased CBV throughout most of the enhancing tumor, with a mean tumor decrease of 22 percent (right column).
The identification of early predictors of clinical endpoints (e.g., overall survival) could reduce the duration and cost of clinical trials. Towards this end, the predictive potential of DSC-MRI in GBM patients was recently evaluated in ACRIN 6677/RTOG 0625, a multicenter, randomized, phase II trial of bevacizumab with irinotecan or temozolomide (36). Changes in tumor CBV before and at 2, 8 and 16 weeks after treatment initiation were correlated with overall survival (OS). Significant decreases in CBV at two weeks were observed in patients with an OS greater than one year, whereas patients with increases in tumor CBV were found to have significantly shorter OS. This trial highlights the potential of CBV as a prognostic biomarker of treatment response in recurrent GBM patients, particularly in the context of therapeutic agents targeting angiogenic pathways.
With the increasing use of DSC-MRI in clinical trials and routine practice there is growing interest in the field to standardize image acquisition and post-processing strategies (37). While there is a general consensus on the most robust acquisition strategies (e.g., pulse sequence type and parameters, CBV quantification, correction techniques for contrast agent leakage effects), current efforts aim to address the challenges of harmonizing these techniques across MRI vendors and data analysis packages.
3.2 Dynamic contrast enhanced MRI (DCE-MRI)
DCE-MRI acquires heavily T1-weighted images before, during, and after injection of a CA leading to an increase in signal intensity on T1-weighted images yielding a time-intensity curve reflecting the delivery and retention of contrast agent within the tissue of interest. DCE-MRI is a class of techniques characterized by whether a qualitative, semi-quantitative, or quantitative approach is used for data analysis. A qualitative analysis examines the shape (e.g., plateau or persistent) of the time-intensity curve (38,39), while a semi-quantitative analysis provides values such as the area under the curve (AUC), enhancement, time to peak, and wash-in/wash-out slopes (38,40). A quantitative analysis fits the time-intensity curve to pharmacokinetic models to extract parameters that reflect physiological characteristics such as tumor vessel perfusion and permeability and tissue volume fractions (40). Although applying quantitative models to the DCE-MRI data is more complex than qualitative or semi-quantitative approaches, the extracted parameters provide (in principle) a more direct measure of vascular characteristics. The Tofts-Kety model is most frequently used and considers the contrast agent distributed between two compartments, the blood/plasma space (Cp) and the tissue space (Ct) (41). In 1999, Tofts et al. standardized quantitative DCE-MRI notation where Ktrans is the volume transfer constant between Cp and Ct, kep is the redistribution rate constant between Ct and Cp, and the plasma and tissue volume fractions are denoted as vp and ve, respectively (42).
DCE-MRI has played a role in the assessment of anticancer therapies, as well as in the prediction of eventual response in a variety of cancers (43–47, and Figure 2). An early report using DCE-MRI as a study endpoint in a Phase I clinical trial was in 2002 when 5,6-dimethylxanthenone-4-acetic acid was used to treat patients with advanced solid tumors (48). Despite the small sample size, significant reductions in the AUC were reported in nine of the 16 patients at 24 hours after the first dose (48). More recently, DCE-MRI was investigated in a Phase I trial of patients with prostate cancer treated with cediranib. In a majority of the patients, Dahut et al. observed rapid and sustained reductions in AUC and Ktrans from baseline up to two or more cycles of therapy (49). Additionally, Ktrans at baseline was associated with progression free survival suggesting that DCE-MRI may also be a predictive biomarker of clinical outcome (49). DCE-MRI was found to be predictive of pathological complete response (pCR) in patients with stage II/III breast cancer undergoing neoadjuvant chemotherapy. Li et al. found that after one cycle of therapy, kep predicted pCR with a sensitivity and specificity of 0.83 and 0.65, respectively (50).
Figure 2.
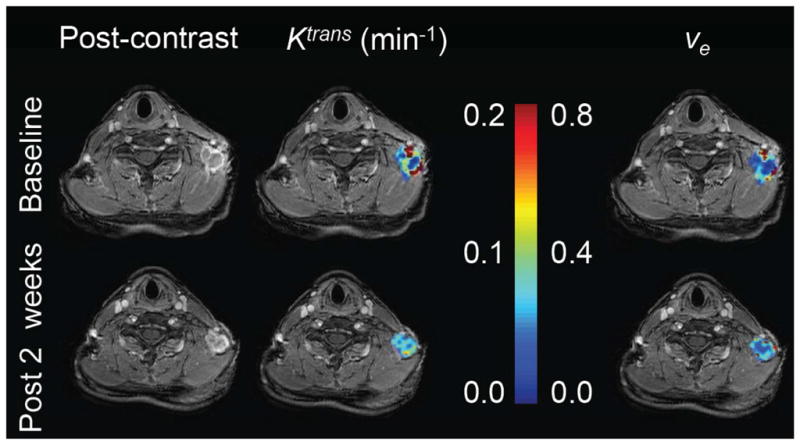
Representative example of DCE-MRI employed in a clinical trial investigating the efficacy of a novel PI3K inhibitor in combination with cisplatin. Quantitative MRI data were collected at baseline and after two weeks of therapy in a patient with metastatic triple-negative breast cancer. Note that there is no appreciable difference in tumor size between imaging time points, however the decrease in Ktrans suggests a decrease vascular perfusion and permeability. ve appears to be unchanged after treatment.
Even with these successes, several limitations of DCE-MRI have been identified (45,51,52) emphasizing the need for its systematic evaluation in assessing treatment response and predicting clinical outcomes. Accuracy and precision of the estimated quantitative parameters can be affected by the estimation of the arterial input function, spatial and temporal resolutions, pharmacokinetic models, and curve fitting strategies. On the subject of model fitting, Huang et al was the first to compare 12 DCE-MRI software tools in a multicenter data analysis challenge (46). Ktrans, kep, ve, and vp from 10 patients before and after the first cycle of NAC were analyzed using site-specific models and algorithms. Although considerable parameter variations were observed, agreement in parameter percentage change was better than that in absolute parameters. Further systematic evaluations assessing reproducibility, evaluating efficacy in a specific patient population and therapy, and finally expanding into a multicenter study are required. Reproducibility studies are important in order to establish the range outside of which any observed changes would be due to therapy and not measurement error (53,54). The reproducibility of several semi-quantitative and quantitative parameters has been investigated in patients with solid tumors (54,55). While there have been some excellent efforts at evaluating semi-quantitative DCE-MRI in a large multicenter trial (see, e.g., 56), more studies are needed before DCE-MRI can be fully utilized in routine clinical care.
3.3 Diffusion weighted imaging (DWI)
In diffusion-weighted imaging (DWI), the image contrast reflects the distance water molecules can migrate or “diffuse” from their original spatial position over a short time interval due to random, thermally induced motion (i.e., Brownian motion). By acquiring two or more images with different degrees of “diffusion weighting” (obtained by applying the diffusion sensitizing gradients with different amplitudes on successive image acquisitions), an estimate of the amount of molecular water diffusion, termed the apparent diffusion coefficient (ADC), can be calculated at each voxel using the equation
| [1] |
where S is the signal intensity measured with application of a diffusion-sensitizing gradient, S0 is the signal intensity with no diffusion-sensitizing gradient, and b is a composite variable reflecting various acquisition parameters (including the strength of the gradient pulse, duration of the pulse, and interval between pulses) (57). For a more extensive review of the physics of DWI, the reader is referred to (58).
Cancers often exhibit significantly reduced ADC values when compared to healthy tissues, a finding typically attributed to the increased cell density of many malignancies (59). With treatment, intratumoral ADC values typically rise, presumably because of decreases in cell density consequent to apoptosis and cell death, with concomitant disruption of cell membranes allowing water molecules to diffuse more freely. This basic paradigm—low tumor ADC values before treatment followed by rising tumor ADC values with effective treatment—provides the basic model for DWI as a technique for response assessment (please see Figure 3 for an illustrative example).
Figure 3.
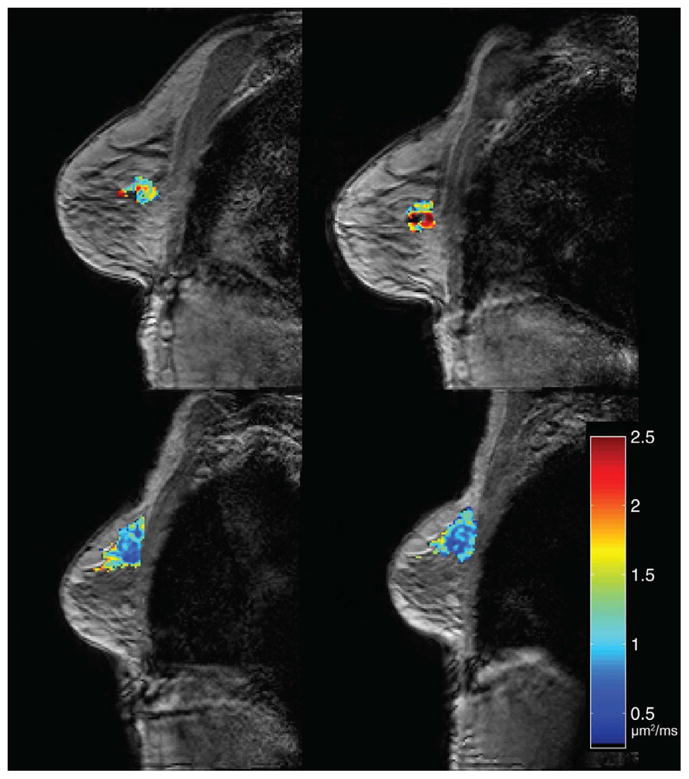
Changes in ADC in response to neoadjuvant therapy in breast cancer can be measured early in the course of therapy. Shown here are examples of ADC maps acquired before the start of therapy (left column) and after one cycle of therapy (right column) for a patient who went on to have a pathological complete response (top row) and a patient who did not respond (bottom row). (ADC values are shown in units of μm2/ms.)
One of the potential advantages of DWI for evaluating treatment response over standard response criteria, such as RECIST, is that it is sensitive to changes occurring at the cellular level prior to changes in gross tumor size. Recent studies have demonstrated changes in ADC after a single cycle of neoadjuvant treatment for breast cancer and these changes correlate with pathological outcome (60,61). Changes in ADC one month after transcatheter arterial chemoembolization (TACE) were predictive of progression free survival in hepatocelluar carcinoma (HCC) (62). DWI also provides a means of evaluating the response of anti-angiogenic drugs. For example, studies of patients receiving bevacizumab for newly diagnosed (63) and for recurrent glioblastoma (64) both demonstrated that characteristics of the tumor ADC histograms at early time points in treatment may be useful for determining patient outcome.
Even with these promising results, there still remain several challenges that must be overcome before DWI is routinely used in the clinical setting. The standard image acquisition techniques used to acquire DWIs are susceptible to image artifacts (58). In the ACRIN 6677/RTOG 0625 trial (65), only 47% of the 123 patients had high quality diffusion data free of image distortion and only 68% were considered usable. The complex physiological factors that affect ADC measurements are also a limitation. It is generally assumed that the measured ADC primarily reflects tumor cellularity; however, there are several biological processes (e.g., edema and perfusion) that can affect ADC values. Ellingson et al. (65) hypothesized that the increase in ADC they measured in patients who sown early disease progression was an indicator that the drug was not effectively reducing vascular edema rather than a change in tumor cellularity. Data analysis methods must be validated as well. Analyses using mean tumor ADC alone may not be able to predict patient response as well as more advanced analysis methods, such as functional diffusion mapping (fDM) (60) or multiparametric analyses (61; see section 4.4 below).
In summary, DWI is a valuable tool for quantitative imaging and treatment assessment, relying only on endogenous contrast mechanisms. It can be applied in a variety of applications and disease sites. Future work includes standardizing protocols, improving image quality, and performing additional multi-center trials.
4. Emerging MRI Methods for Cancer
4.1 Chemical exchange saturation transfer MRI
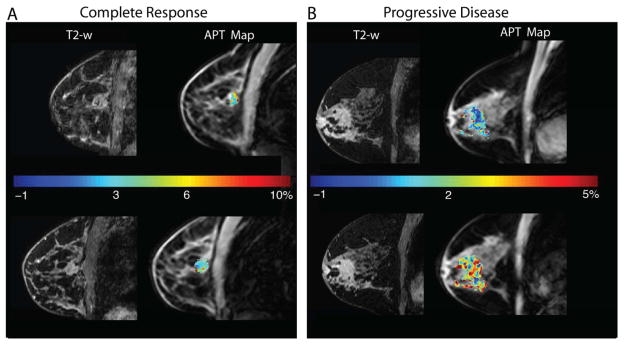
Chemical exchange saturation transfer (CEST) is a technique enabling indirect detection of tissue metabolites via exchangeable protons. The exchangeable protons that resonate at a frequency distinct from bulk water protons are selectively saturated via many off-resonance (with respect to water protons) pulses prior to imaging (66). The saturated species are thought to interact with the magnetization of the bulk water through direct chemical exchange, which reduces the observed water signal. Of particular interest for cancer imaging is the amide proton transfer (APT) metric (see Figure 4), reflective of the concentration of amide protons and their exchange rate with the free proton pool (67). This APT metric has been used to assess physical and physiological characteristics of the tissue microenvironment such as temperature, pH, and metabolite concentration (68–70).
Figure 4.
The figure displays APT maps in breast cancer patients who underwent MRI examination at 3 T before (top row) and after a single cycle (bottom row) of neoadjuvant therapy. Panel A shows T2-weighted images on the left and APT maps on the right for a patient who achieved a pathological complete response at the end of therapy. Panel B shows similar results for a patient that had residual disease at the conclusion of neoadjuvant therapy. The mean APT values decreased by 27% for the responder (Panel A) while this metric increased by 78% for the non-responder (Panel B).
A z-spectrum, which is the measured water signal, S(Δω), normalized by the signal without saturation (S0) plotted as a function the offset frequency (Δω) of the saturating irradiation, is used to assess the CEST effects present in a tissue (71). The z-spectrum is characterized by a symmetric direct saturation around the water frequency (Δω = 0 ppm) and aberrations from this symmetry at the resonances of the exchangeable protons, particularly that due to APT (Δω = 3.5 ppm). These asymmetries are quantified via magnetization transfer ratio (MTR) asymmetry analysis (72) calculated by subtracting the right (−Δω) and left (Δω) signal intensity ratios:
| [2] |
which can be used to examine the z-spectra asymmetry caused by the APT (Δω = 3.5 ppm), termed APTasym:
| [3] |
The APTasym was initially applied in humans to assess amide proton content (thought to be proportional to mobile protein or peptides; 68), and their exchange rate (thought to be reflective of tissue pH) (68, 70, 73) in brain tumors at 3T (74). This study, as well as those following (75) including migration to 7T (76,77) demonstrate that APTasym is increased in glioma relative to surrounding tissue. This increase in APT contrast is hypothesized to be a result of tumor cells accumulating defective proteins at a higher rate than normal while also experiencing alterations in pH due to hypoxia (78). Contrary to magnetic resonance spectroscopy, CEST MRI has sufficient sensitivity to allow imaging due to the signal enhancement, which facilitates clinical translation. Preclinical (79) and clinical (74,80) studies indicate the ability to both distinguish tumor from edema as well as perform tumor grading (80,81).
APTasym is a unique contrast offering complementary information to that provided by standard clinical MRI measures; however, it is not without limitations. For example, CEST imaging in vivo is a complex technique because of interferences with direct water saturation (spillover effect; 82), the involvement of other exchanging pools (83), in particular macromolecular systems (magnetization transfer, MT; 84), and nuclear Overhauser effects (NOEs, 85). Moreover, there is a strong dependence of the measured effects on the sequence parameters of radiofrequency irradiation for selective saturation which makes the comparison of results obtained at different laboratories difficult (86).
4.2 Hyperpolarized MRI
MR hyperpolarization technology allows increasing nuclear spin polarization to the order unity (or 100%) significantly above the equilibrium P level; thus, the process of hyperpolarization enables unprecedented MRI sensitivity gains by >10,000 fold, which is achieved through transient manipulation with the agent molecule. The hyperpolarized substrate molecule can be administered via i.v. injection or inhalation (typically as a bolus) into a living organism (87). There are multiple biomedical hyperpolarization technologies that have already demonstrated their potential in humans (87,88): dissolution Dynamic Nuclear Polarization (d-DNP) (89), Parahydrogen Induced Polarization (PHIP) (90,91), Signal Amplification by Reversible Exchange (SABRE) (92) and Spin Exchange Optical Pumping (SEOP) (93). The main goal of the hyperpolarization process is to produce a sufficiently large batch of hyperpolarized contrast agent (HCA) with sufficiently long lifetime (i.e., long T1) for its administration and in vivo distribution and subsequent metabolism. As a result, most HCA include a low-gamma heteronucleus (129Xe, 13C, 15N, 3He, etc.) used for hyperpolarization storage and detection (87), because protons typically have low T1 values on the order of a few seconds, though there are exceptions when long-lived states of protons are employed such as those in hyperpolarized propane gas (94,95). HCAs are non-radioactive, and they can report on both uptake and metabolism, because it is possible to discern multiple metabolites and report on their distribution (114) using difference in chemical shifts (97) or in J-couplings of multiple metabolites (115). Moreover, HCAs’ T1 and lifetimes are typically within minutes, and therefore, HCAs signals are quickly cleared, and multiple administrations of HCAs can be conducted within the same imaging session. Furthermore, a hyperpolarized MRI scan requires only a few seconds (111,116). In addition, the detection sensitivity of hyperpolarized MRI does not depend on B0 of the main scanner (117), and high-quality images can be potentially obtained with low-field MRI (i.e. ≤ 0.3T), which can have a significantly lower costs and greater patient throughput than high-field MRI. High-resolution human images were reported at magnetic field strengths of 0.2 T with hyperpolarized 129Xe (118) and 0.007 T with hyperpolarized 3He (119).
HCAs can be successfully used for quantitative imaging (please see Box 3). For example, the ratio of injected hyperpolarized 13C-pyruvate to produced 13C-lactate in tumors is correlated with the aggressiveness of prostate cancer (120), and the hyperpolarized 13C-lactate intensity is correlated with response to treatment (98). The ratio of injected hyperpolarized bicarbonate and produced hyperpolarized CO2 can directly report on pH (104). However, hyperpolarized MRI has a major shortcoming in that the actual produced signal is proportional to the product of P and metabolite concentration, and the exact knowledge of the concentration is hindered by the differential T1 relaxation processes of multiple metabolites (e.g., 13C-pyruvate and 13C-lactate pair) in multiple compartments (e.g., relaxation in blood and in tumor). These challenges can be potentially overcome with the use of a single metabolite: for example, the use of hyperpolarized 15N-heterocycles produced by d-DNP (121) and SABRE (122) technologies can be used for pH sensing, where the chemical shift itself of the molecular probe is highly sensitive to the pH environment (121) – with potential application to cancer imaging, because acidic pH is frequently a property of cancer (123).
Box 3. HCAs for quantitative imaging.
In less than 20 years (96), hyperpolarization technologies enabled validation of many HCAs in animal models of human diseases including the use of 13C-pyruvate (97–99), 13C-lactate (100), 13C-glucose (101), 13C-fructose (102), 13C-succinate (103), 13C-fumarate (104), 13C-glutamine (105) in cancer imaging, 13C-bicarbonate (106) for pH imaging, 13C-tetrafluoropropionate (107) for plaque imaging, 129Xe and 3He for lung imaging (108), 129Xe for brown fat imaging (109) among others (87,110). Moreover, hyperpolarized 129Xe, 3He, and 13C-pyruvate have already been successfully tested in clinical trials (111–113).
Despite the above advantages of hyperpolarized MRI, there are two major translational barriers. First, the preparation of HCAs requires (frequently expensive) isotopic labeling, and expensive hyperpolarization equipment with relatively low throughput. Second, most HCA molecules have a heteronuclear hyperpolarized site (e.g., 13C or 129Xe) requiring multinuclear MRI scanner capability – a feature not widely available on MRI scanners. These fundamental challenges can be potentially solved through the use of less expensive hyperpolarization techniques (e.g., SABRE vs. d-DNP) or through innovation in hyperpolarization hardware (116) or through the invention of HCAs with long-lived proton sites vs. heteronuclear-based HCAs (e.g., hyperpolarized 1H-propane vs. hyperpolarized 129Xe). Moreover, heteronuclear-based HCAs can also potentially be detected via indirect proton detection (38); the latter would require a relatively minor clinical MRI scanner upgrade, and would therefore enable this technology on most clinical MRI scanners.
4.3 Magnetic Resonance Elastography
The fundamental link between tissue mechanics and disease has led to the development of technologies for quantitative assessment of mechanical stiffness in tissue through non-invasive imaging, termed “elastography” (125–132). A primary motivation for the use of elastography in cancer response assessment and prediction is based on direct evidence linking the progression of cancerous tissue to the disruption and concurrent stiffening of the stromal extracellular matrix structural architecture (133–136). Elevated interstitial fluid pressure within tumors also contributes to observations of elevated stiffness and correlates with cancer progression and therapeutic resistance (137). Many new cancer therapeutics seek to directly target the abnormal cancer niche (138), including drugs with specific anti-fibrotic activity (139). Thus, it is of great import to develop imaging based methods to provide a noninvasive measure of the mechanical stiffness of the tissue extracellular matrix.
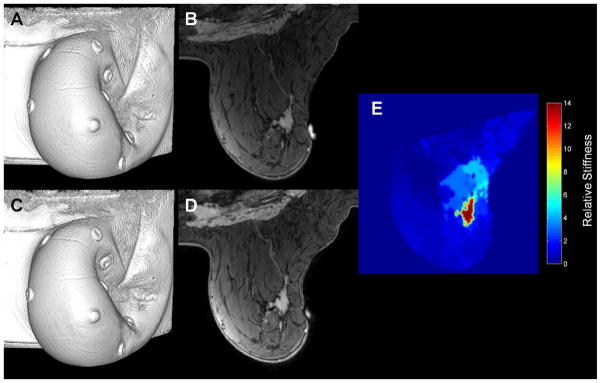
As a general method, elastography involves applying mechanical excitation, imaging the displacement response, and computing spatial estimates of tissue mechanical elasticity. While first demonstrated using ultrasound (125), elastography has been applied in many imaging modalities, including magnetic resonance, computed tomography, and optical imaging. Magnetic resonance elastography (MRE), in particular, allows for quantitative evaluation of tissue mechanical stiffness over a large field of view and deep within the body (140). Mechanical excitation, either dynamic or quasi-static, is typically applied externally by coupling to an acoustic, piezoelectric, or pneumatic deformation source. In the dynamic case, tissue response to mechanical excitation is typically visualized using phase-contrast imaging and motion sensitive pulse sequences synchronized to the frequency of applied excitation (140). In the quasi-static case, image volumes are acquired before and after the application of mechanical deformation (141,142). Quantitative estimates of tissue stiffness are then calculated based on the observed tissue displacement and an assumed material constitutive relationship (typically linear elasticity), through direct inversion or biomechanical model-based methods. An example of quasi-static MRE with biomechanical model-based reconstruction of tissue stiffness as applied to breast cancer assessment is shown in Figure 5.
Figure 5.
An example of quasi-static MR elastography in breast cancer. Image volumes and central slice images before (panels A and B) and after (panels C and D) the application of an external mechanical deformation are used, along with a biomechanical model, to estimate the tissue mechanical stiffness (panel E). Cancer is typically revealed to be significantly stiffer than surrounding healthy tissue.
Preliminary applications of MRE in the clinic has been made for assessing hepatic fibrosis, and is rapidly emerging as a successful non-invasive image-based alternative to percutaneous tissue biopsy (131). While the number of MRE studies in cancer is limited, recent investigations have begun to show promise for the use of MRE in characterization of this disease. For, example, Venkatesh et al. used MRE to show that mechanical stiffness could differentiate malignant focal liver lesions from benign lesions, normal liver tissue, and fibrotic liver tissue (143). In this preliminary study, malignant liver lesions were found to be significantly stiffer than benign lesions (10.1 kPa versus 2.7 kPa, p < 0.001) with 100% accuracy. MRE for lesion characterization has also shown promise in breast (144,145,146) and prostate (147) cancers. Challenging the simplifying assumptions of linear biomechanical constitutive relationships, Garteiser et al. used a viscoelastic mechanical model and extracted estimates of the storage modulus (elasticity component) and the loss modulus (viscous component), and found a significant elevation in the viscous component of the viscoelastic MRE signal in malignant breast tumors as compared to benign (148).
While MRE has recently shown promise for response assessment in several pre-clinical cancer studies (149,150,151), significantly more work needs to be performed in order to advance MRE for use in clinical therapy response assessment and prediction. Many more patients in this setting must be examined with MRE in order to evaluate the predictive performance. Additionally, methodological advancements will be necessary to address the limited spatial resolution and signal quality of traditional MRE examinations. These technical challenges must be overcome for robust longitudinal response assessment for small lesions and within-lesion heterogeneity. Finally, correlations of MRE with histopathology will be important for further understanding of the biological basis of these examinations.
4.4 Multi-parameter MRI methods
As indicated above, quantitative MRI techniques are playing an increasingly important role in oncology for detecting lesions, monitoring therapy, or predicting treatment response. A relatively new approach to increase the accuracy of tumor identification or prediction of therapy response is to integrate the data available from multi-parametric MRI. The general hypothesis is that combining the (potentially) complementary information on tumor properties available from multiple MRI measures will increase the ability to detect, monitor, and predict outcome. For example, there have been many studies showing that multi-parameter MRI can achieve this goal in prostate cancer (152–156). In the study by Turkbey et al (152), 70 patients with biopsy-proved prostate cancer with a median Gleason score of 7, were imaged by T2-weighted MRI, DCE-MRI, and magnetic resonance spectroscopy (MRS). On T2-weighted images, the criterion to detect prostate cancer was a well-circumscribed, round-ellipsoid low intensity lesion. On MRS, the criterion for identifying tumor tissue was a choline-citrate ratio ≥ 3 standard deviations above the mean value of healthy tissue. On DCE-MRI, tumor location was evaluated by visual interpretation as well as Ktrans and kep parametric maps. The results showed that the combination of T2, DCE-MRI, and MRS increased the probability of tumor detection from approximately 0.38 (DCE-MRI alone) to 0.78 (combining all measures).
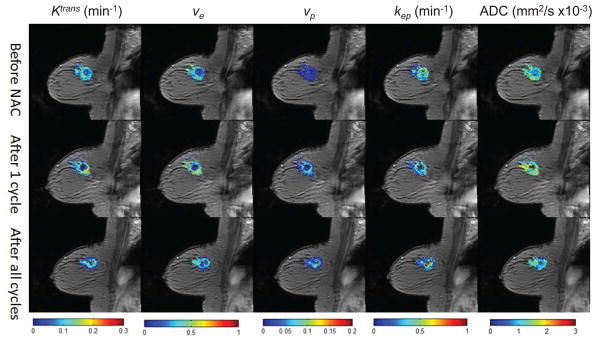
There have also been efforts investigating multi-parameter MRI methods for assessing or predicting the response of breast tumors to neoadjuvant chemotherapy, and several recent publications have demonstrated that combining multiple parameters improves predictive ability (157–163). (See Figure 6 for an illustrative example.) In the study by Hylton et al. (164), 216 patients with invasive breast cancer of ≥ 3 cm were imaged by MR at four time points: before NAC, after one cycle of anthracyline-based treatment, between the anthracycline-based regimen and taxane, and after all cycles of NAC. The longest diameter of the primary tumor, tumor volume, signal enhancement ratio (SER) at MR imaging, and clinical tumor size were assessed and changes in each parameter from baseline to each time point were fit to a univariate random-effects logistic regression model to predict predict pathological complete response and residual cancer burden (RCB). A multivariate model was also performed and adjusted for race and age. Higher areas under receiver operating characteristic curve (AUCs) were found for longest diameter and tumor volume than for clinical size at all the time points in the univariate analysis. When all four variables were considered in the multivariate analysis, the AUCs for predicting pathological complete response increased to 0.75 and 0.84, at the early time point and prior to surgery, respectively. Similarly, the AUCs for predicting residual cancer burden also increased to 0.71 and 0.81.
Figure 6.
Parametric maps of Ktrans, ve, vp, kep, and ADC are displayed before (top row), after 1 cycle (middle row), and after all cycles (bottom row) of neoadjuvant chemotherapy for a patient that had residual tumor burden. It is clear that each one of these maps displays their own spatial variations and report on different aspects of tumor status. Thus, combining them to increase (for example) the predictive value of quantitative MRI is a natural line of investigation.
Two relatively new areas of multi-parametric imaging include using such data to initialize and constrain predictive mechanistic models of tumor growth and treatment response (see, e.g., (165) and references cited therein), and to relate tumor phenotype to genomic signatures (166). Both of these approaches have seen much interest in recent years though with many technical improvements and initial applications in patients. However, much work is required to bring these methods to routine application in clinical trials. More broadly, while multi-parametric methods are gaining more attention in oncology, consensus on the optimal practice in image acquisition, data processing, and interpretation has yet to be determined and is an active area of investigation. Indeed, for each of the above MRI methods described above, we have tried to list current shortcomings of the techniques and these issues are only compounded when two (or more) methods are combined.
5. Summary
Quantitative MRI in oncology had undergone enormous advances in the last decade with a number of techniques now routinely used in clinical trials. Furthermore, there are a number of methods that are rapidly evolving and have shown early promise in preliminary clinical studies. Going forward, it is imperative that consensus among data acquisition and analysis methods is achieved and repeatability and reproducibility is established so that quantitative MRI can be intelligently applied for particular disease types and therapeutic regiments.
KEY POINTS.
The fundamental limitations of RECIST must be addressed by more quantitative imaging methods; MRI offers a number of existing and emerging methods to fill this need.
DCE-MRI, DSC-MRI, and diffusion MRI have all advanced to the point where they can offer quantitative insights into tumor characteristics and these techniques are now frequently employed in clinical trials either alone or in concert.
MR elastography, CEST, and hyperpolarized MRI are three emerging techniques that can offer insights complimentary to those provided by the diffusion and perfusion MRI methods.
The ability to acquire multiple data types in a single MRI session provides the opportunity to combine these methods in a multi-parametric approach, which has been shown to have increased clinical value over single parameter methods.
SYNOPSIS.
We discuss eight areas of quantitative MRI that are either currently employed (RECIST, DCE-MRI, DSC-MRI, diffusion MRI) in clinical trials, or are emerging (CEST, elastography, hyperpolarized MRI, multi-parameter MRI) as promising techniques in diagnosing cancer and assessing or predicting response of cancer to therapy. After a brief introduction of the technique, we summarize illustrative applications of the technique in the clinical setting before describing the current limitations of the methods.
Acknowledgments
We thank the National Institutes of Health, the National Science Foundation, and the Department of Defense for funding through NCI U01CA142565, NCI R25CA092043, NCI U01CA174706, NCI R01CA158079, NCATS KL2 RR024977, NSF CHE-1416268, NIH 1R21EB018014, and the DOD CDMRP breast cancer award W81XWH-12-1-0159/BC112431.
Footnotes
DISCLOSURE STATEMENT
The Authors have nothing to disclose
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Richard G. Abramson, Email: Richard.g.abramson@vanderbilt.edu, Institute of Imaging Science, Department of Radiological and Radiological Sciences, Vanderbilt University
Lori Arlinghaus, Email: lori.arlinghaus@Vanderbilt.Edu, Department of Radiological and Radiological Sciences, Vanderbilt University
Adrienne Dula, Email: adrienne.dula@Vanderbilt.Edu, Institute of Imaging Science, Department of Radiological and Radiological Sciences, Vanderbilt University
C. Chad Quarles, Email: chad.quarles@Vanderbilt.Edu, Institute of Imaging Science, Departments of Radiological and Radiological Sciences, Biomedical Engineering, Cancer Biology, Vanderbilt University
Ashley Stokes, Email: ashley.m.stokes@Vanderbilt.Edu, Institute of Imaging Science, Department of Radiological and Radiological Sciences, Vanderbilt University
Jared Weis, Email: jared.a.weis@Vanderbilt.Edu, Department of Biomedical Engineering, Vanderbilt University
Jennifer Whisenant, Email: j.whisenant@Vanderbilt.Edu, Institute of Imaging Science, Department of Radiological and Radiological Sciences, Vanderbilt University
Eduard Y. Chekmenev, Email: eduard.chekmenev@Vanderbilt.Edu, Institute of Imaging Science, Departments of Radiological and Radiological Sciences, Biomedical Engineering, and Biochemistry, Vanderbilt University
Igor Zhukov, Email: i.zhukov@inbox.ru, National Research Nuclear University, MEPhI, Moscow 115409, Russia
Jason Williams, Email: jason.m.williams@Vanderbilt.Edu, Institute of Imaging Science, Vanderbilt University
Thomas Yankeelov, Email: thomas.yankeelov@vanderbilt.edu, Institute of Imaging Science, Departments of Radiological and Radiological Sciences, Biomedical Engineering, Physics, and Cancer Biology, Vanderbilt University
References
- 1.Buckler AJ, Bresolin L, Dunnick NR, et al. A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiology. 2011;258(3):906–14. doi: 10.1148/radiol.10100799. [DOI] [PubMed] [Google Scholar]
- 2.Rosenkrantz AB, Mendiratta-Lala M, Bartholmai BJ, et al. Clinical utility of quantitative imaging. Acad Radiol. 2015;22(1):33–49. doi: 10.1016/j.acra.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yankeelov TE, Abramson RG, Quarles CC. Quantitative multimodality imaging in cancer research and therapy. Nat Rev Clin Oncol. 2014;11(11):670–80. doi: 10.1038/nrclinonc.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gore JC, Manning HC, Quarles CC, et al. Magnetic resonance in the era of molecular imaging of cancer. Magn Reson Imaging. 2011;29(5):587–600. doi: 10.1016/j.mri.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahl RL, Jacene H, Kasamon Y, et al. From recist to percist: Evolving considerations for pet response criteria in solid tumors. J Nucl Med. 2009;50(Suppl 1):122S–50S. doi: 10.2967/jnumed.108.057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurland BF, Gerstner ER, Mountz JM, et al. Promise and pitfalls of quantitative imaging in oncology clinical trials. Magn Reson Imaging. 2012;30(9):1301–12. doi: 10.1016/j.mri.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Micheel C, Ball J Institute of Medicine (U.S.). Committee on Qualification of Biomarkers and Surrogate Endpoints in Chronic Disease. Evaluation of biomarkers and surrogate endpoints in chronic disease. Washington, D.C: National Academies Press; 2010. pp. 97–116. [PubMed] [Google Scholar]
- 8.Abramson RG, Burton KR, Yu JP, et al. Methods and challenges in quantitative imaging biomarker development. Acad Radiol. 2015;22(1):25–32. doi: 10.1016/j.acra.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke LP, Nordstrom RJ, Zhang H, et al. The quantitative imaging network: Nci’s historical perspective and planned goals. Transl Oncol. 2014;7(1):1–4. doi: 10.1593/tlo.13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comis RL, Schnall MD. Opportunities for the ECOG-ACRIN cancer research group within the new national clinical trials network. Semin Oncol. 2015;42(1):1–3. doi: 10.1053/j.seminoncol.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi-Sekino S, Sekino M, Ueno S. Biological effects of electromagnetic fields and recently updated safety guidelines for strong static magnetic fields. Magn Reson Med Sci. 2011;10(1):1–10. doi: 10.2463/mrms.10.1. [DOI] [PubMed] [Google Scholar]
- 12.Chakeres DW, Bornstein R, Kangarlu A. Randomized comparison of cognitive function in humans at 0 and 8 tesla. J Magn Reson Imaging. 2003;18(3):342–5. doi: 10.1002/jmri.10366. [DOI] [PubMed] [Google Scholar]
- 13.Chakeres DW, de Vocht F. Static magnetic field effects on human subjects related to magnetic resonance imaging systems. Prog Biophys Mol Biol. 2005;87(2–3):255–65. doi: 10.1016/j.pbiomolbio.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Kangarlu A, Burgess RE, Zhu H, et al. Cognitive, cardiac, and physiological safety studies in ultra high field magnetic resonance imaging. Magn Reson Imaging. 1999;17(10):1407–16. doi: 10.1016/s0730-725x(99)00086-7. [DOI] [PubMed] [Google Scholar]
- 15.Raunig DL, McShane LM, Pennello G, et al. Quantitative imaging biomarkers: A review of statistical methods for technical performance assessment. Stat Methods Med Res. 2015;24(1):27–67. doi: 10.1177/0962280214537344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shellock FG, Spinazzi A. Mri safety update 2008: Part 2, screening patients for mri. AJR Am J Roentgenol. 2008;191(4):1140–9. doi: 10.2214/AJR.08.1038.2. [DOI] [PubMed] [Google Scholar]
- 17.Abramson RG, McGhee CR, Lakomkin N, Arteaga CL. Pitfalls in RECIST Data Extraction for Clinical Trials: Beyond the Basics. Academic radiology. 2015;22(6):779–786. doi: 10.1016/j.acra.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) European journal of cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer. 1981;47(1):207–214. doi: 10.1002/1097-0142(19810101)47:1<207::aid-cncr2820470134>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 20.Jain RK, Lee JJ, Ng C, et al. Change in tumor size by RECIST correlates linearly with overall survival in phase I oncology studies. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30(21):2684–2690. doi: 10.1200/JCO.2011.36.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buyse M, Thirion P, Carlson RW, et al. Relation between tumour response to first-line chemotherapy and survival in advanced colorectal cancer: a meta-analysis. Meta-Analysis Group in Cancer Lancet. 2000;356(9227):373–378. doi: 10.1016/s0140-6736(00)02528-9. [DOI] [PubMed] [Google Scholar]
- 22.Pazdur R. Response rates, survival, and chemotherapy trials. Journal of the National Cancer Institute. 2000;92(19):1552–1553. doi: 10.1093/jnci/92.19.1552. [DOI] [PubMed] [Google Scholar]
- 23.Paesmans M, Sculier JP, Libert P, et al. Response to chemotherapy has predictive value for further survival of patients with advanced non-small cell lung cancer: 10 years experience of the European Lung Cancer Working Party. European journal of cancer. 1997;33(14):2326–2332. doi: 10.1016/s0959-8049(97)00325-0. [DOI] [PubMed] [Google Scholar]
- 24.Aronen HJ, Cohen MS, Belliveau JW, et al. Ultrafast imaging of brain tumors. Topics in Magnetic Resonance Imaging. 1993;5(1):14–24. [PubMed] [Google Scholar]
- 25.Aronen HJ, Gazit IE, Louis DN, et al. Cerebral blood volume maps of gliomas: comparison with tumor grade and histologic findings. Radiology. 1994;191(1):41–51. doi: 10.1148/radiology.191.1.8134596. [DOI] [PubMed] [Google Scholar]
- 26.Ostergaard L, Weisskoff RM, Chesler DA, et al. High resolution measurement of cerebral blood flow using intravascular tracer bolus passages. Part I: Mathematical approach and statistical analysis. Magnetic Resonance in Medicine. 1996;36(5):715–25. doi: 10.1002/mrm.1910360510. [DOI] [PubMed] [Google Scholar]
- 27.Donahue KM, Krouwer HG, Rand SD, et al. Utility of simultaneously acquired gradient-echo and spin-echo cerebral blood volume and morphology maps in brain tumor patients. Magnetic Resonance in Medicine. 2000;43(6):845–53. doi: 10.1002/1522-2594(200006)43:6<845::aid-mrm10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 28.Schmainda KM, Rand SD, Joseph AM, et al. Characterization of a first-pass gradient-echo spin-echo method to predict brain tumor grade and angiogenesis. AJNR Am J Neuroradiol. 2004;25(9):1524–32. [PMC free article] [PubMed] [Google Scholar]
- 29.Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol. 2006;27(4):859–67. [PMC free article] [PubMed] [Google Scholar]
- 30.Law M, Yang S, Wang H, Babb JS, Johnson G, Cha S, Knopp EA, Zagzag D. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR: American Journal of Neuroradiology. 2003;24(10):1989–98. [PMC free article] [PubMed] [Google Scholar]
- 31.Weber MA, Zoubaa S, Schlieter M, et al. Diagnostic performance of spectroscopic and perfusion MRI for distinction of brain tumors. Neurology. 2006;66(12):1899–906. doi: 10.1212/01.wnl.0000219767.49705.9c. [DOI] [PubMed] [Google Scholar]
- 32.Law M, Oh S, Babb JS, et al. Low-grade gliomas: dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging--prediction of patient clinical response. Radiology. 2006;238(2):658–67. doi: 10.1148/radiol.2382042180. [DOI] [PubMed] [Google Scholar]
- 33.Boxerman JL, Ellingson BM, Jeyapalan S, et al. Longitudinal DSC-MRI for Distinguishing Tumor Recurrence From Pseudoprogression in Patients With a High-grade Glioma. American journal of clinical oncology. 2014 doi: 10.1097/COC.0000000000000156. [DOI] [PubMed] [Google Scholar]
- 34.Gahramanov S, Raslan AM, Muldoon LL, et al. Potential for differentiation of pseudoprogression from true tumor progression with dynamic susceptibility-weighted contrast-enhanced magnetic resonance imaging using ferumoxytol vs. gadoteridol: a pilot study. Int J Radiat Oncol Biol Phys. 2011;79(2):514–23. doi: 10.1016/j.ijrobp.2009.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsien C, Galban CJ, Chenevert TL, et al. Parametric response map as an imaging biomarker to distinguish progression from pseudoprogression in high-grade glioma. J Clin Oncol. 2010;28(13):2293–9. doi: 10.1200/JCO.2009.25.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmainda KM, Zhang Z, Prah M, et al. Dynamic susceptibility contrast MRI measures of relative cerebral blood volume as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 multicenter trial. Neuro-oncology. 2015 doi: 10.1093/neuonc/nou364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welker K, Boxerman J, Kalnin A, et al. ASFNR Recommendations for Clinical Performance of MR Dynamic Susceptibility Contrast Perfusion Imaging of the Brain. AJNR Am J Neuroradiol. 2015;36(6):E41–51. doi: 10.3174/ajnr.A4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnes SL, Whisenant JG, Loveless ME, et al. Practical Dynamic Contrast Enhanced MRI in Small Animal Models of Cancer: Data Acquisition, Data Analysis, and Interpretation. Pharmaceutics. 2012;4(3):442–78. doi: 10.3390/pharmaceutics4030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhl CK, Mielcareck P, Klaschik S, et al. Dynamic breast MR imaging: are signal intensity time course data useful for differential diagnosis of enhancing lesions? Radiology. 1999;211(1):101–10. doi: 10.1148/radiology.211.1.r99ap38101. [DOI] [PubMed] [Google Scholar]
- 40.Yankeelov TE, Gore JC. Dynamic Contrast Enhanced Magnetic Resonance Imaging in Oncology: Theory, Data Acquisition, Analysis, and Examples. Curr Med Imaging Rev. 2009;3(2):91–107. doi: 10.2174/157340507780619179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kety SS. The theory and applications of the exchange of inert gas at the lungs and tissues. Pharmacol Rev. 1951;3(1):1–41. [PubMed] [Google Scholar]
- 42.Tofts PS1, Brix G, Buckley DL, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10(3):223–32. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 43.Padhani AR, Hayes C, Assersohn L, et al. Prediction of clinicopathologic response of breast cancer to primary chemotherapy at contrast-enhanced MR imaging: initial clinical results. Radiology. 2006;239(2):361–74. doi: 10.1148/radiol.2392021099. [DOI] [PubMed] [Google Scholar]
- 44.Johansen R, Jensen LR, Rydland J, et al. Predicting survival and early clinical response to primary chemotherapy for patients with locally advanced breast cancer using DCE-MRI. J Magn Reson Imaging. 2009;29(6):1300–7. doi: 10.1002/jmri.21778. [DOI] [PubMed] [Google Scholar]
- 45.Hahn OM, Yang C, Medved M, et al. Dynamic contrast-enhanced magnetic resonance imaging pharmacodynamic biomarker study of sorafenib in metastatic renal carcinoma. J Clin Oncol. 2008;26(28):4572–8. doi: 10.1200/JCO.2007.15.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly RJ, Rajan A, Force J, et al. Evaluation of KRAS mutations, angiogenic biomarkers, and DCE-MRI in patients with advanced non-small-cell lung cancer receiving sorafenib. Clin Cancer Res. 2011;17(5):1190–9. doi: 10.1158/1078-0432.CCR-10-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer JM, Perlewitz KS, Hayden JB, et al. Phase I trial of preoperative chemoradiation plus sorafenib for high-risk extremity soft tissue sarcomas with dynamic contrast-enhanced MRI correlates. Clin Cancer Res. 2013;19(24):6902–11. doi: 10.1158/1078-0432.CCR-13-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galbraith SM, Rustin GJ, Lodge MA, et al. Effects of 5,6-dimethylxanthenone-4-acetic acid on human tumor microcirculation assessed by dynamic contrast-enhanced magnetic resonance imaging. J Clin Oncol. 2002;20(18):3826–40. doi: 10.1200/JCO.2002.09.144. [DOI] [PubMed] [Google Scholar]
- 49.Dahut WL, Madan RA, Karakunnel JJ, et al. Phase II clinical trial of cediranib in patients with metastatic castration-resistant prostate cancer. BJU International. 2013;111(8):1269–80. doi: 10.1111/j.1464-410X.2012.11667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Abramson RG, Arlinghaus LR, et al. Multiparametric magnetic resonance imaging for predicting pathological response after the first cycle of neoadjuvant chemotherapy in breast cancer. Invest Radiol. 2015;50(4):195–204. doi: 10.1097/RLI.0000000000000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim KA, Park MS, Ji HJ, et al. Diffusion and perfusion MRI prediction of progression-free survival in patients with hepatocellular carcinoma treated with concurrent chemoradiotherapy. J Magn Reson Imaging. 2014;39(2):286–92. doi: 10.1002/jmri.24161. [DOI] [PubMed] [Google Scholar]
- 52.Barnes SL, Quarles CC, Yankeelov TE. Modeling the effect of intra-voxel diffusion of contrast agent on the quantitative analysis of dynamic contrast enhanced magnetic resonance imaging. PLoS One. 2014;9(9):e108726. doi: 10.1371/journal.pone.0108726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barnes SL, Whisenant JG, Loveless ME, et al. Assessing the reproducibility of dynamic contrast enhanced magnetic resonance imaging in a murine model of breast cancer. Magn Reson Med. 2013;69(6):1721–34. doi: 10.1002/mrm.24422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galbraith SM, Lodge MA, Taylor NJ, et al. Reproducibility of dynamic contrast-enhanced MRI in human muscle and tumours: comparison of quantitative and semi-quantitative analysis. NMR Biomed. 2002;15(2):132–42. doi: 10.1002/nbm.731. [DOI] [PubMed] [Google Scholar]
- 55.Jackson A, Jayson GC, Li KL, et al. Reproducibility of quantitative dynamic contrast-enhanced MRI in newly presenting glioma. The British journal of radiology. 2003;76(903):153–62. doi: 10.1259/bjr/70653746. [DOI] [PubMed] [Google Scholar]
- 56.Hylton NM, Blume JD, Bernreuter WK, et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy--results from ACRIN 6657/I-SPY TRIAL. Radiology. 2012;263(3):663–72. doi: 10.1148/radiol.12110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le Bihan D, Breton E, Lallemand D, et al. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161(2):401–407. doi: 10.1148/radiology.161.2.3763909. [DOI] [PubMed] [Google Scholar]
- 58.Arlinghaus LR, Yankeelov TE. Diffusion-weighted MRI. In: Yankeelov T, Pickens DR, Price RR, editors. Quantitative MRI in cancer, Imaging in medical diagnosis and therapy. Boca Raton, FL: CRC Press; 2012. pp. 81–97. [Google Scholar]
- 59.Charles-Edwards EM, deSouza NM. Diffusion-weighted magnetic resonance imaging and its application to cancer. Cancer Imaging. 2006;6:135–143. doi: 10.1102/1470-7330.2006.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galban CJ, Ma B, Malyarenko D, et al. Multi-site clinical evaluation of DW-MRI as a treatment response metric for breast cancer patients undergoing neoadjuvant chemotherapy. PloS one. 2015;10(3):e0122151. doi: 10.1371/journal.pone.0122151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X, Abramson RG, Arlinghaus LR, et al. Multiparametric magnetic resonance imaging for predicting pathological response after the first cycle of neoadjuvant chemotherapy in breast cancer. Investigative radiology. 2015;50(4):195–204. doi: 10.1097/RLI.0000000000000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vandecaveye V, Michielsen K, De Keyzer F, et al. Chemoembolization for hepatocellular carcinoma: 1-month response determined with apparent diffusion coefficient is an independent predictor of outcome. Radiology. 2014;270(3):747–757. doi: 10.1148/radiol.13130591. [DOI] [PubMed] [Google Scholar]
- 63.Wen Q, Jalilian L, Lupo JM, et al. Comparison of ADC metrics and their association with outcome for patients with newly diagnosed glioblastoma being treated with radiation therapy, temozolomide, erlotinib and bevacizumab. Journal of neuro-oncology. 2015;121(2):331–339. doi: 10.1007/s11060-014-1636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pope WB, Qiao XJ, Kim HJ, et al. Apparent diffusion coefficient histogram analysis stratifies progression-free and overall survival in patients with recurrent GBM treated with bevacizumab: a multi-center study. Journal of neuro-oncology. 2012;108(3):491–498. doi: 10.1007/s11060-012-0847-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ellingson BM, Kim E, Woodworth DC, et al. Diffusion MRI quality control and functional diffusion map results in ACRIN 6677/RTOG 0625: a multicenter, randomized, phase II trial of bevacizumab and chemotherapy in recurrent glioblastoma. Int J Oncol. 2015;46(5):1883–1892. doi: 10.3892/ijo.2015.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolff SD, Balaban RS. Magnetization transfer contrast (MTC) and tissue water proton relaxation in vivo. Magn Reson Med. 1989;10(1):135–44. doi: 10.1002/mrm.1910100113. [DOI] [PubMed] [Google Scholar]
- 67.van Zijl PC, Zhou J, Mori N, et al. Mechanism of magnetization transfer during on-resonance water saturation. A new approach to detect mobile proteins, peptides, and lipids. Magn Reson Med. 2003;49(3):440–9. doi: 10.1002/mrm.10398. [DOI] [PubMed] [Google Scholar]
- 68.Zhou J, Payen JF, Wilson DA, et al. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med. 2003 Aug;9(8):1085–90. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 69.Sun PZ, Zhou J, Sun W, et al. Detection of the ischemic penumbra using pH-weighted MRI. J Cereb Blood Flow Metab. 2007;27(6):1129–36. doi: 10.1038/sj.jcbfm.9600424. [DOI] [PubMed] [Google Scholar]
- 70.Ward KM, Balaban RS. Determination of pH using water protons and chemical exchange dependent saturation transfer (CEST) Magn Reson Med. 2000;44(5):799–802. doi: 10.1002/1522-2594(200011)44:5<799::aid-mrm18>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 71.Bryant RG. The dynamics of water-protein interactions. Annu Rev Biophys Biomol Struct. 1996;25:29–53. doi: 10.1146/annurev.bb.25.060196.000333. [DOI] [PubMed] [Google Scholar]
- 72.Guivel-Scharen V, Sinnwell T, Wolff SD, Balaban RS. Detection of proton chemical exchange between metabolites and water in biological tissues. J Magn Reson. 1998;133(1):36–45. doi: 10.1006/jmre.1998.1440. [DOI] [PubMed] [Google Scholar]
- 73.Sun PZ, Murata Y, Lu J, et al. Relaxation-compensated fast multislice amide proton transfer (APT) imaging of acute ischemic stroke. Magn Reson Med. 2008;59(5):1175–82. doi: 10.1002/mrm.21591. [DOI] [PubMed] [Google Scholar]
- 74.Jones CK, Schlosser MJ, van Zijl PC, et al. Amide proton transfer imaging of human brain tumors at 3T. Magn Reson Med. 2006;56(3):585–92. doi: 10.1002/mrm.20989. [DOI] [PubMed] [Google Scholar]
- 75.Zhou J, Blakeley JO, Hua J. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med. 2008;60(4):842–9. doi: 10.1002/mrm.21712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones CK, Polders D, Hua J, et al. In vivo three-dimensional whole-brain pulsed steady-state chemical exchange saturation transfer at 7 T. Magn Reson Med. 2012;67(6):1579–89. doi: 10.1002/mrm.23141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mougin OE, Coxon RC, Pitiot A, Gowland PA. Magnetization transfer phenomenon in the human brain at 7 T. Neuroimage. 2010;49(1):272–81. doi: 10.1016/j.neuroimage.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 78.Salhotra A, Lal B, Laterra J, et al. Amide proton transfer imaging of 9L gliosarcoma and human glioblastoma xenografts. NMR Biomed. 2008;21(5):489–97. doi: 10.1002/nbm.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou J, Lal B, Wilson DA, et al. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med. 2003;50(6):1120–6. doi: 10.1002/mrm.10651. [DOI] [PubMed] [Google Scholar]
- 80.Zhou J, Blakeley JO, Hua J, et al. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med. 2008;60(4):842–9. doi: 10.1002/mrm.21712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wen Z, Hu S, Huang F, et al. MR imaging of high-grade brain tumors using endogenous protein and peptide-based contrast. Neuroimage. 2010;51(2):616–22. doi: 10.1016/j.neuroimage.2010.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mulkern RV, Williams ML. The general solution to the Bloch equation with constant rf and relaxation terms: application to saturation and slice selection. Med Phys. 1993;20(1):5–13. doi: 10.1118/1.597063. [DOI] [PubMed] [Google Scholar]
- 83.van Zijl PC, Jones CK, Ren J, et al. MRI detection of glycogen in vivo by using chemical exchange saturation transfer imaging (glycoCEST) Proc Natl Acad Sci U S A. 2007;104(11):4359–64. doi: 10.1073/pnas.0700281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Desmond KL, Stanisz GJ. Understanding quantitative pulsed CEST in the presence of MT. Magn Reson Med. 2012;67(4):979–90. doi: 10.1002/mrm.23074. [DOI] [PubMed] [Google Scholar]
- 85.Li H, Zu Z, Zaiss M, et al. Imaging of amide proton transfer and nuclear Overhauser enhancement in ischemic stroke with corrections for competing effects. NMR Biomed. 2015;28(2):200–9. doi: 10.1002/nbm.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun PZ, van Zijl PC, Zhou J. Optimization of the irradiation power in chemical exchange dependent saturation transfer experiments. J Magn Reson. 2005;175(2):193–200. doi: 10.1016/j.jmr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 87.Kurhanewicz J, Vigneron DB, Brindle K, et al. Analysis of Cancer Metabolism by Imaging Hyperpolarized Nuclei: Prospects for Translation to Clinical Research Neoplasia. 2011;13(2):81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nikolaou P, Goodson BM, Chekmenev EY. NMR Hyperpolarization Techniques for Biomedicine. Chem Eur J. 2015;21(8):3156–3166. doi: 10.1002/chem.201405253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ardenkjaer-Larsen JH, Fridlund B, Gram A, et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100(18):10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bowers CR, Weitekamp DP. Transformation of Symmetrization Order to Nuclear-Spin Magnetization by Chemical-Reaction and Nuclear-Magnetic-Resonance. Phys Rev Lett. 1986;57(21):2645–2648. doi: 10.1103/PhysRevLett.57.2645. [DOI] [PubMed] [Google Scholar]
- 91.Eisenschmid TC, Kirss RU, Deutsch PP, et al. Para Hydrogen Induced Polarization In Hydrogenation Reactions. J Am Chem Soc. 1987;109(26):8089–8091. [Google Scholar]
- 92.Adams RW, Aguilar JA, Atkinson KD, et al. Reversible Interactions with para-Hydrogen Enhance NMR Sensitivity by Polarization Transfer. Science. 2009;323(5922):1708–1711. doi: 10.1126/science.1168877. [DOI] [PubMed] [Google Scholar]
- 93.Walker TG, Happer W. Spin-exchange optical pumping of noble-gas nuclei. Rev Mod Phys. 1997;69(2):629–642. [Google Scholar]
- 94.Kovtunov KV, Truong ML, Barskiy DA, et al. Long-lived Spin States for Low-field Hyperpolarized Gas MRI. Chem Eur J. 2014;20(45):14629–14632. doi: 10.1002/chem.201405063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kovtunov KV, Truong ML, Barskiy DL, et al. Propane-d6 Heterogeneously Hyperpolarized by Parahydrogen. J Phys Chem C. 2014;118(48):28234–28243. doi: 10.1021/jp508719n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Golman K, Axelsson O, Johannesson H, et al. Parahydrogen-induced polarization in imaging: Subsecond C-13 angiography. Magn Reson Med. 2001;46(1):1–5. doi: 10.1002/mrm.1152. [DOI] [PubMed] [Google Scholar]
- 97.Golman K, in’t Zandt R, Thaning M. Real-time metabolic imaging. Proc Natl Acad Sci U S A. 2006;103(30):11270–11275. doi: 10.1073/pnas.0601319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Day SE, Kettunen MI, Gallagher FA, et al. Detecting tumor response to treatment using hyperpolarized C-13 magnetic resonance imaging and spectroscopy. Nat Med. 2007;13(11):1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 99.Park I, Bok R, Ozawa T, et al. Detection of Early Response to Temozolomide Treatment in Brain Tumors Using Hyperpolarized (13)C MR Metabolic Imaging. J Magn Reson Imaging. 2011;33(6):1284–1290. doi: 10.1002/jmri.22563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen AP, Kurhanewicz J, Bok R, et al. Feasibility of using hyperpolarized [1-C-13]lactate as a substrate for in vivo metabolic C-13 MRSI studies. Magnetic Resonance Imaging. 2008;26(6):721–726. doi: 10.1016/j.mri.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rodrigues TB, Serrao EM, Kennedy BWC, et al. Magnetic resonance imaging of tumor glycolysis using hyperpolarized 13C-labeled glucose. Nat Med. 2014;20(1):93–97. doi: 10.1038/nm.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Keshari KR, Wilson DM, Chen AP, et al. Hyperpolarized 2-C-13 -Fructose: A Hemiketal DNP Substrate for In Vivo Metabolic Imaging. J Am Chem Soc. 2009;131(48):17591–17596. doi: 10.1021/ja9049355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zacharias NM, Chan HR, Sailasuta N, et al. Real-Time Molecular Imaging of Tricarboxylic Acid Cycle Metabolism in Vivo by Hyperpolarized 1-C-13 Diethyl Succinate. J Am Chem Soc. 2012;134(2):934–943. doi: 10.1021/ja2040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gallagher FA, Kettunen MI, Hu DE, et al. Production of hyperpolarized [1,4-C-13(2)]malate from [1,4-C-13(2)]fumarate is a marker of cell necrosis and treatment response in tumors. Proc Natl Acad Sci U S A. 2009;106(47):19801–19806. doi: 10.1073/pnas.0911447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gallagher FA, Kettunen MI, Day SE, et al. C-13 MR spectroscopy measurements of glutaminase activity in human hepatocellular carcinoma cells using hyperpolarized C-13-labeled glutamine. Magn Reson Med. 2008;60(2):253–257. doi: 10.1002/mrm.21650. [DOI] [PubMed] [Google Scholar]
- 106.Gallagher FA, Kettunen MI, Day SE, et al. Magnetic resonance imaging of pH in vivo using hyperpolarized C-13-labelled bicarbonate. Nature. 2008;453(7197):940–U973. doi: 10.1038/nature07017. [DOI] [PubMed] [Google Scholar]
- 107.Bhattacharya P, Chekmenev EY, Reynolds WF, et al. Parahydrogen-induced polarization (PHIP) hyperpolarized MR receptor imaging in vivo: a pilot study of 13C imaging of atheroma in mice. NMR Biomed. 2011;24(8):1023–1028. doi: 10.1002/nbm.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Patz S, Hersman FW, Muradian I, et al. Hyperpolarized Xe-129 MRI: A viable functional lung imaging modality? Eur J Radiol. 2007;64(3):335–344. doi: 10.1016/j.ejrad.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Branca RT, He T, Zhang L, et al. Detection of brown adipose tissue and thermogenic activity in mice by hyperpolarized xenon MRI. Proceedings of the National Academy of Sciences. 2014;111(50):18001–18006. doi: 10.1073/pnas.1403697111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brindle KM. Imaging Metabolism with Hyperpolarized 13C-Labeled Cell Substrates. J Am Chem Soc. 2015;137(20):6418–6427. doi: 10.1021/jacs.5b03300. [DOI] [PubMed] [Google Scholar]
- 111.Mugler JP, Altes TA. Hyperpolarized 129Xe MRI of the human lung. J Magn Reson Imaging. 2013;37(2):313–331. doi: 10.1002/jmri.23844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nelson SJ, Kurhanewicz J, Vigneron DB, et al. Metabolic Imaging of Patients with Prostate Cancer Using Hyperpolarized 1-C-13 Pyruvate. Sci Transl Med. 2013;5(198):198ra108. doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Conradi MS, Saam BT, Yablonskiy DA, Woods JC. Hyperpolarized He-3 and perfluorocarbon gas diffusion MRI of lungs. Prog Nucl Magn Reson Spectrosc. 2006;48(1):63–83. [Google Scholar]
- 114.Larson PEZ, Hu S, Lustig M, Kerr AB, et al. Fast Dynamic 3D MR Spectroscopic Imaging With Compressed Sensing and Multiband Excitation Pulses for Hyperpolarized C-13 Studies. Magn Reson Med. 2011;65(3):610–619. doi: 10.1002/mrm.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kennedy BWC, Kettunen MI, Hu D-E, Brindle KM. Probing Lactate Dehydrogenase Activity in Tumors by Measuring Hydrogen/Deuterium Exchange in Hyperpolarized L-[1-C-13, U-H-2]Lactate. J Am Chem Soc. 2012;134(10):4969–4977. doi: 10.1021/ja300222e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nikolaou P, Coffey AM, Walkup LL, et al. Near-unity nuclear polarization with an ‘open-source’ 129Xe hyperpolarizer for NMR and MRI. Proc Natl Acad Sci U S A. 2013;110(35):14150–14155. doi: 10.1073/pnas.1306586110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Coffey AM, Truong ML, Chekmenev EY. Low-field MRI can be more sensitive than high-field MRI. J Magn Reson. 2013;237:169–174. doi: 10.1016/j.jmr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Patz S, Muradian I, Hrovat MI, et al. Human pulmonary imaging and spectroscopy with hyperpolarized Xe-129 at 0. 2T. Acad Radiol. 2008;15(6):713–727. doi: 10.1016/j.acra.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tsai LL, Mair RW, Rosen MS, et al. An open-access, very-low-field MRI system for posture-dependent He-3 human lung imaging. J Magn Reson. 2008;193(2):274–285. doi: 10.1016/j.jmr.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lupo JM, Chen AP, Zierhut ML, et al. Analysis of hyperpolarized dynamic C-13 lactate imaging in a transgenic mouse model of prostate cancer. Magnetic Resonance Imaging. 2010;28(2):153–162. doi: 10.1016/j.mri.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jiang W, Lumata L, Chen W, et al. Hyperpolarized 15N-pyridine Derivatives as pH-Sensitive MRI Agents. Sci Rep. 2015;5:9104. doi: 10.1038/srep09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shchepin RV, Truong ML, Theis T, et al. Hyperpolarization of “Neat” Liquids by NMR Signal Amplification by Reversible Exchange. The Journal of Physical Chemistry Letters. 2015;6(10):1961–1967. doi: 10.1021/acs.jpclett.5b00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 124.Truong ML, Coffey AM, Shchepin RV, et al. Sub-second Proton Imaging of 13C Hyperpolarized Contrast Agents in Water. Contrast Media Mol Imaging. 2014;9(5):333– 341. doi: 10.1002/cmmi.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ophir J, Cespedes I, Ponnekanti H, et al. Elastography - a Quantitative Method for Imaging the Elasticity of Biological Tissues. Ultrasonic Imaging. 1991;13(2):111–134. doi: 10.1177/016173469101300201. [DOI] [PubMed] [Google Scholar]
- 126.Muthupillai R, Lomas DJ, Rossman PJ, et al. Magnetic-Resonance Elastography by Direct Visualization of Propagating Acoustic Strain Waves. Science. 1995;269(5232):1854–1857. doi: 10.1126/science.7569924. [DOI] [PubMed] [Google Scholar]
- 127.Rose GH, Dresner MA, Rossman PJ, et al. “Palpation of the brain” using magnetic resonance elastography. Radiology. 1998;209P:425–425. [Google Scholar]
- 128.Dresner MA, Rose GH, Rossman PJ, et al. Magnetic resonance elastography of the prostate. Radiology. 1998;209P:181–181. [Google Scholar]
- 129.Kallel F, Price RE, Konofagou E, Ophir J. Elastographic imaging of the normal canine prostate in vitro. Ultrasonic Imaging. 1999;21(3):201–215. doi: 10.1177/016173469902100304. [DOI] [PubMed] [Google Scholar]
- 130.Sandrin L, Fourquet B, Hasquenoph JM, et al. Transient elastography: A new noninvasive method for assessment of hepatic fibrosis. Ultrasound in Medicine and Biology. 2003;29(12):1705–1713. doi: 10.1016/j.ultrasmedbio.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 131.Yin M, Talwalkar JA, Glaser KJ, et al. Assessment of hepatic fibrosis with magnetic resonance elastography. Clinical Gastroenterology and Hepatology. 2007;5(10):1207–1213. doi: 10.1016/j.cgh.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Barr RG, Destounis S, Lackey LB, 2nd, et al. Evaluation of breast lesions using sonographic elasticity imaging: a multicenter trial. J Ultrasound Med. 2012;31(2):281–287. doi: 10.7863/jum.2012.31.2.281. [DOI] [PubMed] [Google Scholar]
- 133.Paszek MJ, Weaver VM. The tension mounts: Mechanics meets morphogenesis and malignancy. Journal of Mammary Gland Biology and Neoplasia. 2004;9(4):325–342. doi: 10.1007/s10911-004-1404-x. [DOI] [PubMed] [Google Scholar]
- 134.Paszek MJ, Zahir N, Johnson KR, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 135.Paszek MJ, Zahir N, Lakins JN, et al. Mechano-signaling in mammary morphogenesis and tumorigenesis. Molecular Biology of the Cell. 2004;15:241A–241A. [Google Scholar]
- 136.Huang S, Ingber DE. Cell tension, matrix mechanics, and cancer development. Cancer Cell. 2005;8(3):175–176. doi: 10.1016/j.ccr.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 137.Jain RK, Martin JD, Stylianopoulos T. The role of mechanical forces in tumor growth and therapy. Annu Rev Biomed Eng. 2014;16:321–346. doi: 10.1146/annurev-bioeng-071813-105259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jain RK. Normalizing tumor microenvironment to treat cancer: bench to bedside to biomarkers. J Clin Oncol. 2013;31(17):2205–2218. doi: 10.1200/JCO.2012.46.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Diop-Frimpong B, Chauhan VP, Krane S, et al. Losartan inhibits collagen I synthesis and improves the distribution and efficacy of nanotherapeutics in tumors. Proc Natl Acad Sci U S A. 2011;108(7):2909–2914. doi: 10.1073/pnas.1018892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mariappan YK, Glaser KJ, Ehman RL. Magnetic resonance elastography: a review. Clin Anat. 2010;23(5):497–511. doi: 10.1002/ca.21006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miga MI. A new approach to elastography using mutual information and finite elements. Phys Med Biol. 2003;48(4):467–480. doi: 10.1088/0031-9155/48/4/304. [DOI] [PubMed] [Google Scholar]
- 142.Weis JA, Miga MI, Arlinghaus LR, et al. A mechanically coupled reaction-diffusion model for predicting the response of breast tumors to neoadjuvant chemotherapy. Phys Med Biol. 2013;58(17):5851–66. doi: 10.1088/0031-9155/58/17/5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Venkatesh SK, Yin M, Glockner JF, et al. MR elastography of liver tumors: preliminary results. AJR Am J Roentgenol. 2008;190(6):1534–1540. doi: 10.2214/AJR.07.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.McKnight AL, Kugel JL, Rossman PJ, et al. MR elastography of breast cancer: Preliminary results. American Journal of Roentgenology. 2002;178(6) doi: 10.2214/ajr.178.6.1781411. [DOI] [PubMed] [Google Scholar]
- 145.Siegmann KC, Xydeas T, Sinkus R, et al. Diagnostic value of MR elastography in addition to contrast-enhanced MR imaging of the breast-initial clinical results. European Radiology. 2010;20(2):318–325. doi: 10.1007/s00330-009-1566-4. [DOI] [PubMed] [Google Scholar]
- 146.Weis JA, Miga MI, Li X, et al. Predicting the Response of Breast Cancer to Neoadjuvant Chemotherapy Using a Mechanically Coupled Reaction-Diffusion Model. Cancer Research. doi: 10.1158/0008-5472.CAN-14-2945. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sahebjavaher RS, Nir G, Honarvar M, et al. MR elastography of prostate cancer: quantitative comparison with histopathology and repeatability of methods. NMR Biomed. 2015;28(1):124–139. doi: 10.1002/nbm.3218. [DOI] [PubMed] [Google Scholar]
- 148.Garteiser P, Doblas S, Daire JL, et al. MR elastography of liver tumours: value of viscoelastic properties for tumour characterisation. Eur Radiol. 2012;22(10):2169–2177. doi: 10.1007/s00330-012-2474-6. [DOI] [PubMed] [Google Scholar]
- 149.Pepin KM, Chen J, Glaser KJ, et al. MR elastography derived shear stiffness--a new imaging biomarker for the assessment of early tumor response to chemotherapy. Magn Reson Med. 2014;71(5):1834–1840. doi: 10.1002/mrm.24825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li J, Jamin Y, Boult JK, Cummings C, et al. Tumour biomechanical response to the vascular disrupting agent ZD6126 in vivo assessed by magnetic resonance elastography. Br J Cancer. 2014;110(7):1727–1732. doi: 10.1038/bjc.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weis JA, Flint KM, Sanchez V, et al. Assessing the accuracy and reproducibility of modality independent elastography in a murine model of breast cancer. Journal of Medical Imaging. doi: 10.1117/1.JMI.2.3.036001. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Turkbey B, Pinto P, Mani H, et al. Prostate cancer: value of multiparametric MRI Imaging at 3T for detection – histopathologic correlation. Radiology. 2012;255:89–99. doi: 10.1148/radiol.09090475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Turkbey B, Mani H, Shah V, et al. Multiparametric 3T prostate magnetic resonance imaging to detect cancer: histopathological correlation using prostatectomy specimens processed in customized magnetic resonance imaging based molds. J Urol. 2011;186:1818–24. doi: 10.1016/j.juro.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Delongchamps N, Rouanne M, Flam T, et al. Multiparametric magnetic resonance imaging for the detection and localization of prostate cancer: combination of T2-weighted, dynamic contrast-enhanced and diffusion-weighted imaging. BJU Int. 2010;107:1411–8. doi: 10.1111/j.1464-410X.2010.09808.x. [DOI] [PubMed] [Google Scholar]
- 155.Fütterer JJ, Engelbrecht MR, Huisman HJ, et al. Staging prostate cancer with dynamic contrast-enhanced endorectal MR imaging prior to radical prostatectomy: experienced versus less experienced readers. Radiology. 2005;237(2):541–549. doi: 10.1148/radiol.2372041724. [DOI] [PubMed] [Google Scholar]
- 156.Hoeks CMA, Barentsz JO, Hambrock T, et al. Prostate Cancer: Multiparametric MR Imaging for Detection, Localization, and Staging. Radiology. 2011;261:46–66. doi: 10.1148/radiol.11091822. [DOI] [PubMed] [Google Scholar]
- 157.Yankeelov TE, Lepage M, Chakravarthy A, et al. Integration of quantitative DCEMRI and ADC mapping to monitor treatment response in human breast cancer: initial results. Magn Reson Imaging. 2007;25:1–13. doi: 10.1016/j.mri.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Belli P, Costantini M, Ierardi C, et al. Diffusion-weighted imaging in evaluating the response to neoadjuvant breast cancer treatment. Breast J. 2011;17:610–619. doi: 10.1111/j.1524-4741.2011.01160.x. [DOI] [PubMed] [Google Scholar]
- 159.Li X, Abramson RG, Arlinghaus LR, et al. Multiparametric Magnetic Resonance Imaging for Predicting Pathological Response After the First Cycle of Neoadjuvant Chemotherapy in Breast Cancer. Invest Radiol. 2015;50(4):195–204. doi: 10.1097/RLI.0000000000000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fangberget A, Nilsen LB, Hole KH, et al. Neoadjuvant chemotherapy in breast cancer-response evaluation and prediction of response to treatment using dynamic contrast-enhanced and diffusion-weighted MR imaging. Eur Radiol. 2011;21:1188–1199. doi: 10.1007/s00330-010-2020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hahn SY, Ko EY, Han BK, et al. Role of diffusion-weighted imaging as an adjunct to contrast-enhanced breast MRI in evaluating residual breast cancer following neoadjuvant chemotherapy. Eur J Radiol. 2014;83:283–288. doi: 10.1016/j.ejrad.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 162.Jensen LR, Garzon B, Heldahl MG, et al. Diffusion-weighted and dynamic contrast-enhanced MRI in evaluation of early treatment effects during neoadjuvant chemotherapy in breast cancer patients. J Magn Reson Imaging. 2011;34:1099–1109. doi: 10.1002/jmri.22726. [DOI] [PubMed] [Google Scholar]
- 163.Hylton NM, Blume JD, Bernreuter WK, et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy—results from ACRIN 6657/I-SPY TRIAL. Radiology. 2012;263:663–672. doi: 10.1148/radiol.12110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Huang W, Li X, Chen Y, et al. Variations of Dynamic Contrast-Enhanced Magnetic Resonance Imaging in Evaluation of Breast Cancer Therapy Response: A Multicenter Data Analysis Challenge. Translational Oncology. 2014;7(1):153–166. doi: 10.1593/tlo.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yankeelov TE, Atuegwu N, Hormuth D, et al. Clinically relevant modeling of tumor growth and treatment response. Sci Transl Med. 2013;5(187):187ps9. doi: 10.1126/scitranslmed.3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Colen R, Foster I, Gatenby R, et al. NCI Workshop Report: Clinical and Computational Requirements for Correlating Imaging Phenotypes with Genomics Signatures. Transl Oncol. 2014;7(5):556–69. doi: 10.1016/j.tranon.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]