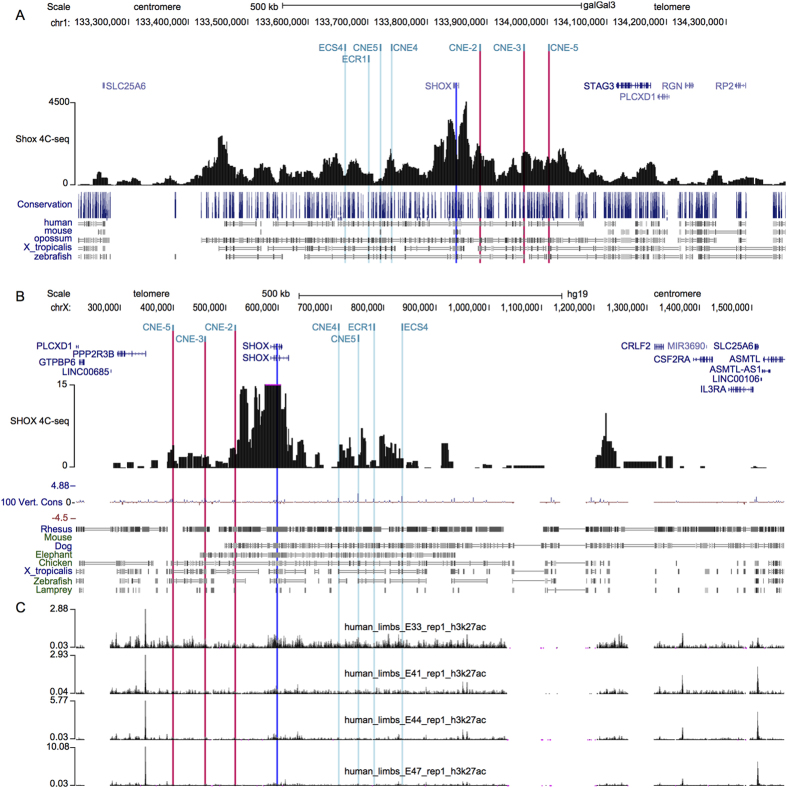
Figure 5. 4C-seq chromatin interaction map of the cis-regulatory region surrounding SHOX.
(A) 4C-seq data of the chicken Shox locus (chr1:133,215,000-134,400,000; UCSC, galGal3) showing that the Shox promoter (blue bar, viewpoint) interacts with 800 kb of adjacent genomic regions that likely contain Shox cis-regulatory regions, a region syntenic with about 1 Mb of human genomic DNA (shown in panel (B)). Red bar marks the position of the upstream CNEs. The 4C-seq data indicate that the Shox regulatory landscape extends both upstream and downstream of the currently known, functionally validated enhancers. (B) 4C-seq data of the human SHOX locus (chrX:215,733–1,561,244; UCSC, hg19) showing that the SHOX promoter (blue bar, viewpoint) interacts with the upstream enhancers (red bars). The RefSeq Genes are shown a the top and the conservation track is included at the bottom of panel (B). (C) Distribution of enhancer-associated H3K27ac histone marks at different human embryonic limb bud stages, produced by Cotney et al. (2013).