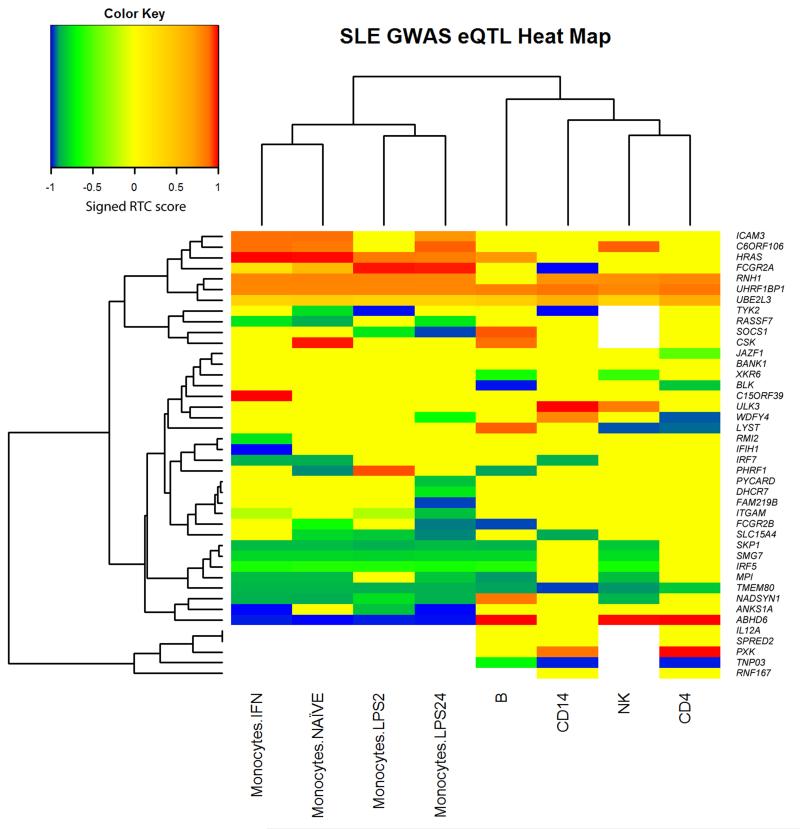
Figure 1.
Heat map for cis-acting gene expression RTC scores from ex vivo cells. The heat map includes all genes with evidence of cis-regulatory (+/– 1Mb) action by SLE associated SNPs in at least one cell type. The color represents a signed-RTC-score: a positive score indicates that the associated allele in the GWAS is positively correlated with gene expression; a negative score indicates that the associated allele in the GWAS is negatively correlated with gene expression. We set the RTC score to zero if the P-value for association was > 0.001. Colors represent the RTC-scores as follows: blue, RTC < –0.9 (GWAS risk allele reduces expression); green, RTC < –0.5 (GWAS risk allele reduces expression); yellow –0.5 < RTC < 0.5; orange, RTC > 0.5 (GWAS risk allele increases expression); red, RTC > 0.9 (GWAS risk allele increases expression). A white block indicates that data were not available for this cell type (see Supplementary Figure 4 for results on lymphoblastoid cell lines), either because the probe data failed QC or the probe was not present in the experiment platform. Clustering was performed on cell types, including only genes with data observed for all cell types (i.e., missing data did not inform cell clustering). Genes were clustered using all available data across cells (missing data were not included when determining distance between pairs of genes if eQTL results were not observed for one of the pairs).